.
S O U R C E : SciTechUpdates
F U L L T E X T P R I M A L S O U R C E : Genome biology (Co-opted transposons help perpetuate conserved higher-order chromosomal structures)
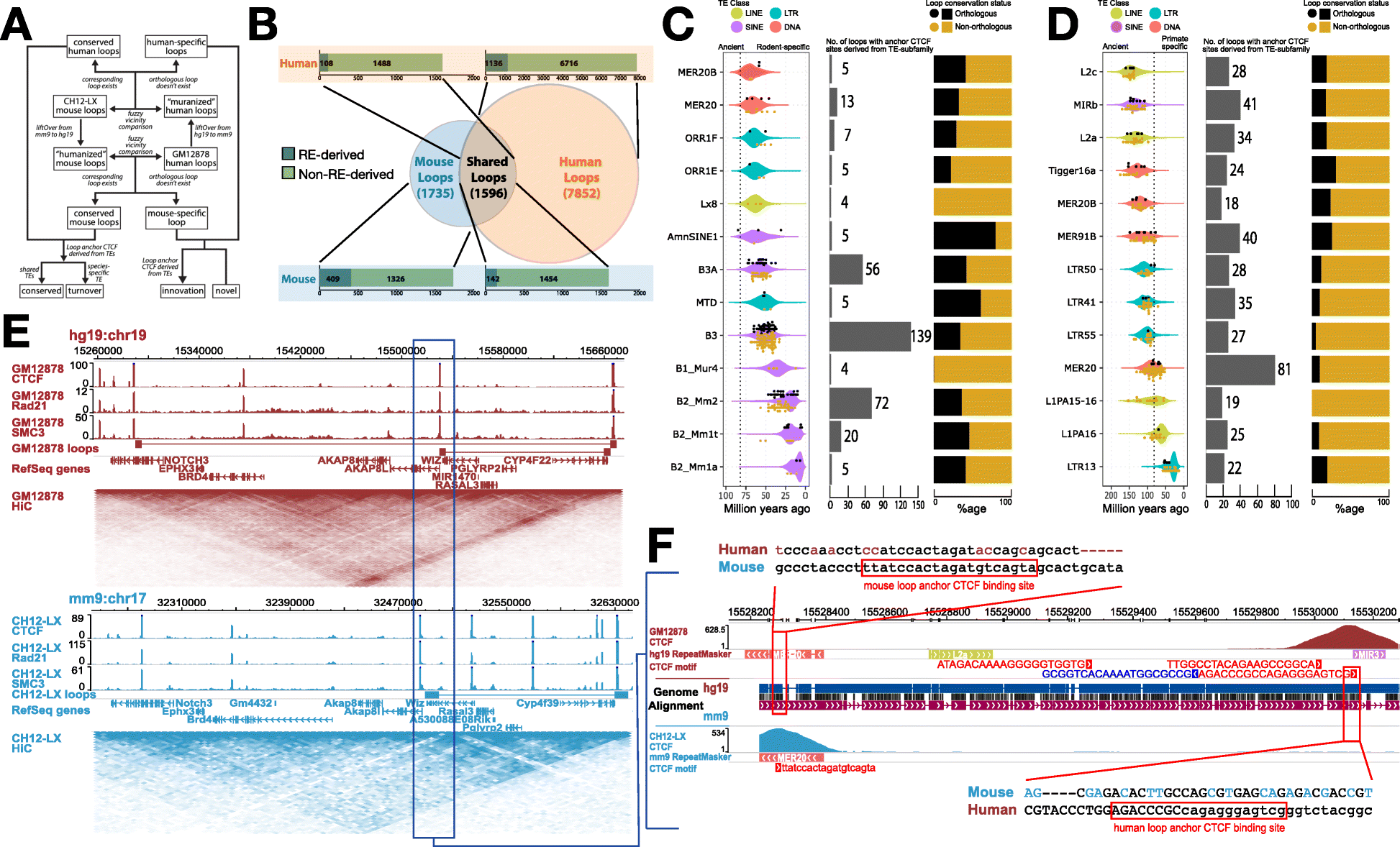
Contribution of TEs to the conservation landscape of human and mouse loops. a Flowchart describing the methodology used to annotate loop orthology. b Venn diagram representing the various classes of chromatin loops based on their orthology and bar plots showing the contribution of REs to anchor CTCFs of each class of loops. c Age distribution and age of individual TEs that contribute loop anchor CTCF sites (black dots for orthologous loops; gold dots for non-orthologous loops) (left), total contribution to loop anchor CTCF sites (middle), distribution of orthologous and non-orthologous loops (right) derived from the top 13 TE subfamilies in mouse and d humans. Estimated primate/rodent divergence time (82 million years ago) is from Meredith et al. [47]. e Contact maps representing a conserved chromatin loop in a syntenic region between human and mouse. f A MER20 transposon insertion provides a redundant CTCF motif that helps in maintaining the conserved 3D structure in mouse via CTCF binding site turnover with remnants of the ancestral CTCF motif, well conserved in most non-rodent mammals (Additional file 1: Figure S2), still seen in the mouse genome.
Long understood as source of novel genetic traits, jumping genes also provide genomic stability. “Jumping genes” – bits of DNA that can move from one spot in the genome to another – are well-known for increasing genetic diversity over the long course of evolution. Now, new research at Washington University School of Medicine in St. Louis indicates that such genes, also called transposable elements, play another, more surprising role: stabilizing the 3D folding patterns of the DNA molecule inside the cell’s nucleus. The study appears Jan. 24 in the journal Genome Biology.
The DNA molecule inside the nucleus of any human cell is more than six feet long. To fit into such a small space, it must fold into precise loops that also govern how genes are turned on or off. It might seem counterintuitive that bits of DNA that randomly move about the genome can provide stability to these folding patterns. Indeed, the discovery contradicts a long-held assumption that the precise order of letters in the DNA sequence always dictates the broader structure of the DNA molecule.
“In places where the larger 3D folding of the genome is the same between mice and humans, you expect the sequence of the letters of the DNA anchoring that shape to be conserved there as well,” said senior author Ting Wang, PhD, the Sanford C. and Karen P. Loewentheil Distinguished Professor of Medicine. “But that’s not what we found, at least not in the portions of the genome that in the past have been called ‘junk DNA.'”
Studying DNA folding in mouse and human blood cells, the researchers found that in many regions where the folding patterns of DNA are conserved through evolution, the genetic sequence of the DNA letters establishing these folds is not. It is ever so slightly displaced. But this changing sequence, a genetic turnover, doesn’t cause problems. Because the structure largely stays the same, the function presumably does, too, so nothing of importance changes.
“We were surprised to find that some young transposable elements serve to maintain old structures,” said first author Mayank N.K. Choudhary, a doctoral student in Wang’s lab. “The specific sequence may be different, but the function stays the same. And we see that this has happened multiple times over the past 80 million years, when the common ancestors of mice and humans first diverged from one another.”
The fact that a new transposable element can insert itself and serve the same role as an existing anchor creates a redundancy in the regulatory portions of the genome – regions of the DNA molecule that determine how and when genes are turned on or off.
According to the researchers, this redundancy makes the genome more resilient. In providing both novelty and stability, jumping genes may help the mammalian genome strike a vital balance — allowing animals the flexibility to adapt to a changing climate, for example, while preserving biological functions required for life, protecting against the DNA damage that is wrought by living and reproducing on Earth over the span of deep time, measured in tens to hundreds of millions of years.
Even so, the researchers were careful to distinguish between portions of the genome that hold genes responsible for producing proteins and the rest of the genome. In genes that code for proteins, the genetic sequence and the structure are both conserved, and this study does not contradict that. However, the new research suggests that jumping genes in the non-protein coding areas of the genome follow different rules of conservation than the protein-coding genes.
.../...
.
Edited by Engadin, 27 January 2020 - 04:03 PM.










































