.
S O U R C E : MedicalXpress
O P E N A C C E S S P R I M A L S O U R C E ( . P D F ) : Cell Report (Genome-wide Profiling Identifies DNA Methylation
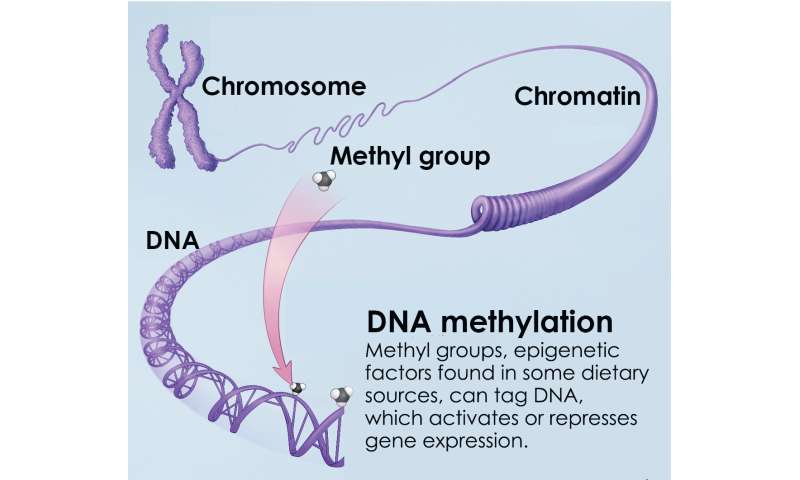
DNA methylation is an epigenetic mechanism essential for normal cell development and differentiation, and is also associated with aging and the formation of cancers. When present, DNA methylation generally represses gene expression. Credit: NIH Common Fund
National Eye Institute (NEI) researchers profiling epigenomic changes in light-sensing mouse photoreceptors have a clearer picture of how age-related eye diseases may be linked to age-related changes in the regulation of gene expression. The findings, published online April 21 in Cell Reports, suggest that the epigenome could be targeted as a therapeutic strategy to prevent leading causes of vision loss, such as age-related macular degeneration (AMD). NEI is part of the National Institutes of Health.
"Our study elucidates the molecular changes and biological pathways linked with aging of rod photoreceptors, light-sensing cells of the retina. Future investigations can now move forward to study how we can prevent or delay vision loss in aging and hopefully reduce the risk of associated neurodegeneration" said the study's lead investigator, Anand Swaroop, Ph.D., senior investigator and chief of the NEI Neurobiology, Neurodegeneration, and Repair Laboratory.
Each organism is born with a genome, a library of genes that control all the body's cellular and tissue functions. Expression of those genes—when information stored in DNA is converted into instructions for making proteins or other molecules—is modulated and maintained by the organism's epigenome. The epigenome tags the DNA code to modify gene expression in ways that can be favorable and unfavorable for survival.
As it turns out, that interplay between the genome and the epigenome evolves as the organism ages. Scientists therefore study epigenomic DNA modifications for clues about why certain diseases develop with advancing age.
To explore how such DNA modifications might influence visual function as we age, Swaroop's team performed whole genome sequencing of DNA methylation changes in mouse rod photoreceptors at four separate stages over the animal's lifetime. DNA methylation is an epigenetic mechanism essential for normal cell development and differentiation, and is also associated with aging and the formation of cancers. When present, DNA methylation generally represses gene expression.
The sequencing was performed at ages three months (young), 12 months (middle-aged), and 18 and 24 months (older). The average lifespan of a mouse is about two years.
Rod photoreceptors are the predominant type of cell in the retina, the light-sensing tissue at the back of the eye. Rod photoreceptors enable dim-light vision, and are critical for the survival of cone photoreceptors that enable daylight and color vision. Rod dysfunction is common in older human adults and can be an early warning sign of AMD and other retinal degenerative diseases.
A simulation shows what the grocery store aisle looks like to someone with age-related macular degeneration. AMD damages the macula, a small spot near the center of the eye's light-sensing retina. The macula lets us see objects that are straight ahead -- it's the part of the eye used for sharp, central vision. For a virtual reality experience, download NEI-VR See What I See at the Apple App store or Google Play. Credit: NEI
The researchers identified 2,054 differentially methylated regions across the four mouse age groups, that is, genomic regions with differences in DNA methylation.
"We know that DNA methylation changes are strongly associated with biological age, but prior to this study we had limited understanding of how these alterations correlated with cellular function," Swaroop said. This is the first study to look at DNA methylation changes as animals age. Very few studies have looked at DNA methylation changes in people with AMD, a leading cause of vision loss in people age 50 and older, which can progress even when vision loss is undetectable.
The researchers then analyzed the differentially methylated regions with RNA sequencing data to look more closely at how the mouse genes were transcribing proteins differently in the retina as the animals aged.
Those analyses uncovered distinct shifts in how the genes produced proteins relevant to energy metabolism, mitochondria function, and the longevity of rod photoreceptors, indicating their contribution to age-related disease susceptibility.
Rod photoreceptors require vast amounts of energy to sustain vision and are thus vulnerable to metabolic stresses that accompany aging. Energy deprivation of photoreceptors is believed to be a key driver of neurodegeneration of the retina.
.../...
.













































