.
O P E N A C C E S S S O U R C E : Microbiome Journal
Abstract
Background
Recent evidence has linked the gut microbiome to host behavior via the gut–brain axis [1,2,3]; however, the underlying mechanisms remain unexplored. Here, we determined the links between host genetics, the gut microbiome and memory using the genetically defined Collaborative Cross (CC) mouse cohort, complemented with microbiome and metabolomic analyses in conventional and germ-free (GF) mice.
Results
A genome-wide association analysis (GWAS) identified 715 of 76,080 single-nucleotide polymorphisms (SNPs) that were significantly associated with short-term memory using the passive avoidance model. The identified SNPs were enriched in genes known to be involved in learning and memory functions. By 16S rRNA gene sequencing of the gut microbial community in the same CC cohort, we identified specific microorganisms that were significantly correlated with longer latencies in our retention test, including a positive correlation with Lactobacillus. Inoculation of GF mice with individual species of Lactobacillus (L. reuteri F275, L. plantarum BDGP2 or L. brevis BDGP6) resulted in significantly improved memory compared to uninoculated or E. coli DH10B inoculated controls. Untargeted metabolomics analysis revealed significantly higher levels of several metabolites, including lactate, in the stools of Lactobacillus-colonized mice, when compared to GF control mice. Moreover, we demonstrate that dietary lactate treatment alone boosted memory in conventional mice. Mechanistically, we show that both inoculation with Lactobacillus or lactate treatment significantly increased the levels of the neurotransmitter, gamma-aminobutyric acid (GABA), in the hippocampus of the mice.
Conclusion
Together, this study provides new evidence for a link between Lactobacillus and memory and our results open possible new avenues for treating memory impairment disorders using specific gut microbial inoculants and/or metabolites.
Background
Specific members of the gut microbiome have been linked to host health and behavior [4]. Intriguingly, probiotics comprised of different Lactobacillus and/or Bifidobacterium strains have been shown to impact behavior in mice, including reduction of symptoms linked to anxiety [5,6,7] and improvement of memory [8, 9]. Administration of probiotic strains specifically results in an improvement in memory of objects and object location [8,9,10,11], but not object temporal order memory [8].
Metabolic clues to memory enhancement have been found by analyzing metabolic signatures in the brains of mice following administration with specific Lactobacillus strains. Increased levels of GABA in the brain is linked to improved working memory and novel object recognition [12, 13]. Mice fed with L. rhamnosus JB-1 had increased mRNA expression of the GABA receptor [5], and increased metabolic levels of GABA in the hippocampus [14]. Increased levels of GABA in the brain could also be due to increased production of GABA by gut bacteria [15, 16]. However, the metabolic mediator(s), if any, between the gut and the brain remain unknown. Recently, O'Hagan et al. (2017) found increased levels of lactate in the brains of mice that were fed supplements containing a mixture of lactobacilli and bifidobacteria: L. acidophilus CUL60, L. acidophilus CUL21, B. bifidum CUL20 and B. lactis CUL34) [8]. Together, these studies suggest a link between specific metabolites produced by lactobacilli and memory of the host via the gut–brain axis that remains to be further explored and validated.
The complex interplay between host genetics, environment, and lifestyle factors and the gut microbiome make studying the role of the microbiome on memory potential difficult in human populations. Model systems can help overcome this barrier and offer many advantages for the study of the genetic basis of complex phenotypes. The “Collaborative Cross” (CC) is a population-based mouse model system with genetic and phenotypic diversity on par with the human population [17]. The CC, which captures nearly 90% of the known variation present in laboratory mice, was established by combining the genomes of eight diverse founder strains (A/J, C57BL/6J, 129S1/SvImJ, NOD/LtJ, NZO/HlLtJ, CAST/EiJ, PWK/PhJ, and WSB/EiJ). The advantage of the CC is that genetic and environmental components of risk can be specified and tightly controlled allowing for a comprehensive analysis of the role of host genetics and the microbiome on memory. In this study for the first time, we performed an unbiased genetic screen using CC mice to identify host genetic and microbiome components that are associated with memory potential. Subsequently, we used this information to focus on specific strains that were correlated with memory in the CC mouse cohort and to evaluate their metabolic profiles in a gnotobiotic mouse system in order to better understand the metabolic mechanisms underlying memory improvement.
Results
We assessed memory using passive avoidance, a fear-motivated test to assess memory-dependent hippocampal function [18, 19], in 535 mice from 29 Collaborative Cross (CC) strains (Table S1). The passive avoidance memory test is based on latency of entry into a compartment where three days earlier, a mild foot shock (0.3 mA for 5 s) was experienced. Mice with good memory avoided entering the chamber where they had previously been exposed to the shock, whereas mice with poor memory entered the chamber. There were significant and reproducible variations in memory potentials across the different CC strains (Fig. 1a). The latency in entry time on the testing day ranged from 87.9 to 600 s (Fig. 1a). Mice from two strains (CC036 and CC010) never entered the chamber within the 600 s assay time. We observed sex differences in memory potential in two of the strains (CC019 and CC032) where memory potential was higher in male mice, whereas no significant sex difference was observed for any of the other strains (Figure S1).
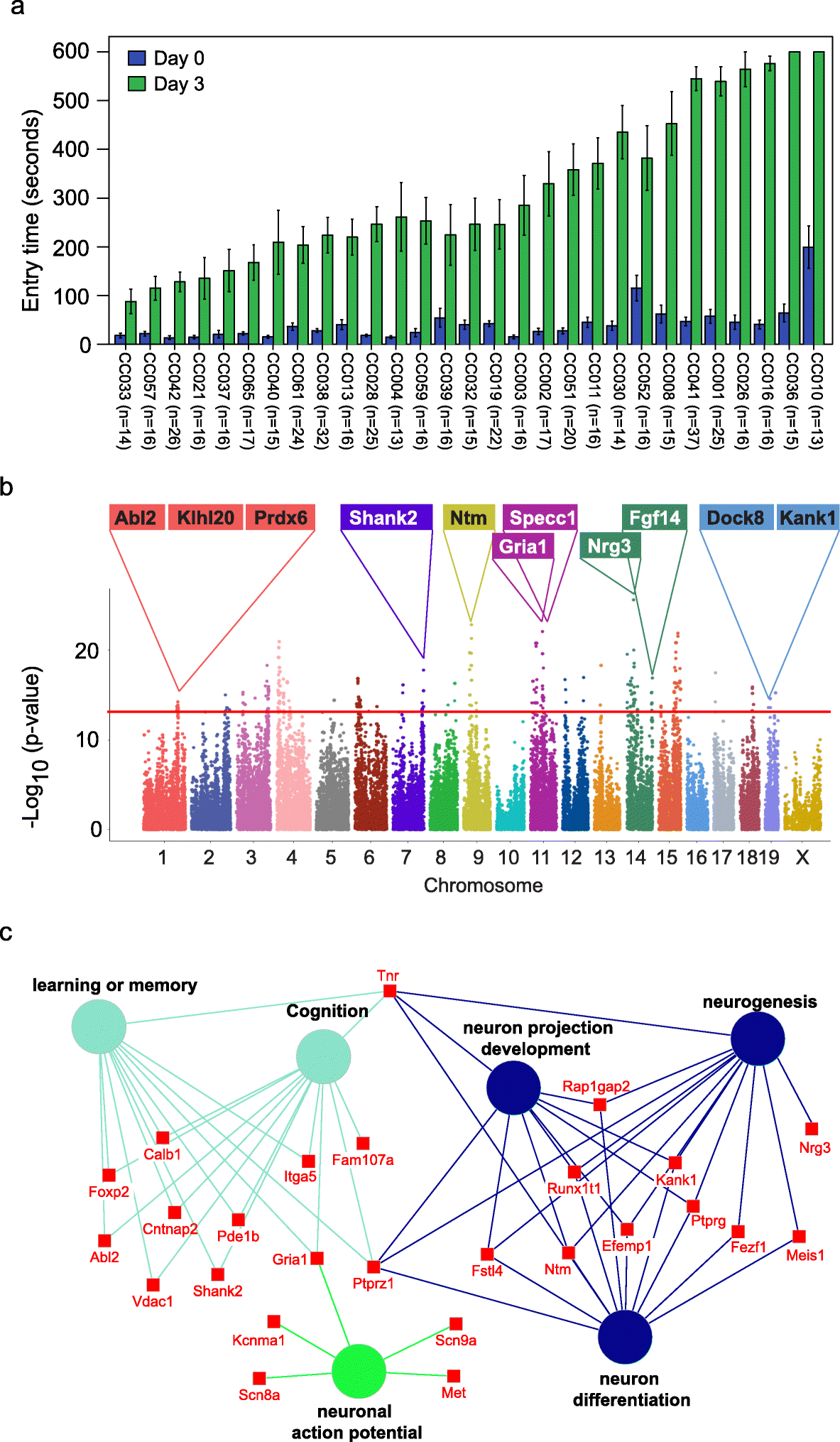
Fig. 1 Identification of genetic variations and candidate genes associated with memory in CC mice.
a Variations in memory across CC strains. Memory was assessed using passive avoidance, a fear-motivated test. The memory test is based on latency of entry into a compartment where 3 days earlier a mild foot shock (0.3 mA for 5 s) was experienced. Entry into the shock compartment on Day 0 is shown in blue, whereas entry 3 days after the foot shock is shown in green (Day 3). Mice with good memory avoided entering the chamber on Day 3, whereas mice with poor memory entered the chamber. Error bars indicate mean ± SEM. b Manhattan plot of the GWAS analysis for memory in CC mice (n = 535 mice). The – log10(P value) is shown for 76,080 SNPs ordered based on genomic position. The horizontal red line indicates the QTL significance threshold at − log10(P value) = 12. Candidate genes previously associated with memory, cognition, or other neurodevelopmental processes located in representative QTL are listed above the plot. c Gene ontology (GO) analysis of genes identified in QTL associated with memory potential in Fig. 1b (n = 535 mice). Genetic loci are significantly enriched for genes implicated in learning or memory, cognition, neuron projection development, neurogenesis, neuron differentiation, and neuronal action potential.
The reproducible variation in memory potential across the CC cohort suggests that host genetics plays an important role in memory. To identify the potential genetic variations that contribute to memory, we performed GWAS with 76080 SNPs across 29 CC strains. We identified 715 SNPs significantly associated with memory (p value < 10−12), corresponding to 222 annotated genes (Fig. 1b, Table S2). Gene set enrichment analysis revealed that the 222 genes were significantly enriched in biological processes related to learning or memory (p = 1.87E–5), neuron cellular component (p = 3.97E–9), and abnormal learning/memory/conditioning phenotypes (p = 1.03E–4; Fig. 1c, Table S3) [20]. In addition to 71 genes known to be associated with memory and learning, our screen also identified 135 genes not previously associated with memory including 65 genes that show expression in the brain based on in situ hybridization data from the Mouse Brain Atlas (Allen Brain Atlas; Table S4). The spatial gene expression data suggest that these 65 genes may play a role in memory.
Previously, we demonstrated natural host variation in the gut microbiome composition across CC mice [21]. To determine links between specific members of the gut microbiome and memory, we correlated 16S rRNA gene sequence data to memory of individual CC strains. Sequence reads were mapped to 5761 OTUs corresponding to 72 bacterial families (Table S5). After filtering OTUs to those with > 100 reads 41 families remained. Four families (Lactobacillaceae, Deferribacteraceae, Bacteroidaceae, and Clostridiaceae) were significantly correlated with memory based on multivariate Cox regression analysis (p < 0.05; Fig. 2a). The hazard ratio (HR) indicated that higher relative abundances of Lactobacillaceae (specifically L. reuteri; HR = 0.79) and Deferribacteraceae (HR = 0.73) and lower relative abundances of Bacteroidaceae (HR = 1.32) and Clostridaceae (HR = 1.32) predicted improved memory potential (Fig. 2a). Because other Lactobacillus strains have previously been implicated in improving memory in mice, humans, and rats: e.g., L. rhamnosus JB-1 in mice [5], L. casei LC122 in aged mice [22], L. helveticus ROO52 in humans [6], and L. acidophilus strains CUL60 and CUL21 (in combination with bifidobacteria) in rats [8]. We focused our remaining studies on Lactobacillus, in particular L. reuteri that was identified in this study.
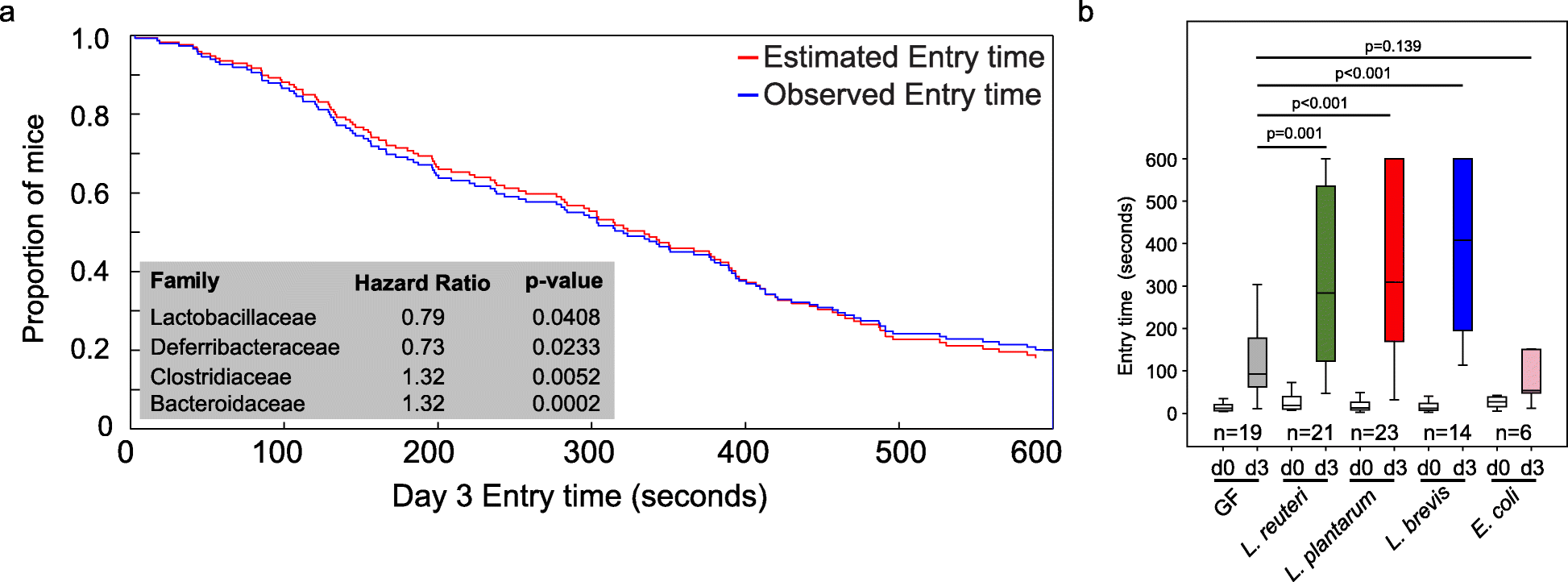
Fig. 2 The impact of Lactobacillus or lactate treatment on memory.
a Identification of microbes associated with memory in CC mice by multivariate Cox regression analysis. b Inoculation of GF mice with individual species of Lactobacillus (L. reuteri F275, L. brevis BDGP6 or L. plantarum BDGP2) resulted in significantly improved memory compared to uninoculated or E. coli inoculated controls. Error bars indicate mean ± SEM
To investigate the impact of Lactobacillus on memory, we colonized separate cohorts of germ-free (GF) mice by oral gavage with L. reuteri F275. This species has an exact 16S rRNA gene sequence match to the OTU that was significantly correlated with improved memory. In addition, we included two other Lactobacillus species for comparison (L. plantarum BDGP2 and L. brevis BDGP6). Within our three inoculated groups, we validated the presence of the individual Lactobacillus OTUs, each of which was dominant in a single treatment group and could be traced to our inoculated species (Figure S2). Memory was assessed using the same passive avoidance test in the Lactobacillus inoculated mice and compared to GF mice of the same genetic background. Eschericia coli DH10B inoculated mice were included as a negative control. Our data showed that all of the Lactobacillus mono-associated mice showed a significant improvement in memory compared to GF mice (p < 0.001; Fig. 2b). In contrast, we observed no memory improvement following application of Escherichia coli DH10B (Fig. 2b).
To identify metabolites produced by lactobacilli that are candidates for microbiome mediated memory enhancement, we assessed the metabolome in fecal samples collected from GF and mice mono-associated with one of each of the three Lactobacillus species: L. reuteri F275, L. plantarum BDGP2 or L. brevis BDGP6 (Fig. 3a, Figure S3, Table S6). The LLE (local-linear embedding analysis) plot shows that the metabolite composition in fecal samples from the Lactobacillus-colonized mice and GF mice were distinct (p = 0.00021; Fig. 3b,c), demonstrating that Lactobacillus inoculation significantly affected the gut metabolome. Based on gas chromatography–mass spectrometry (GC–MS) peak intensities, lactate and threitol were consistently higher in stool samples collected from mice that were colonized with any of the three strains (p < 0.01; Fig. 3d,e; Fig. S3), whereas some strain-specific differences in gut metabolites were seen, such as D-mannitol that was only higher in L. reuteri inoculated samples (p < 0.001; Fig. 3f; Fig. S3). Subsequent injection of mice with mannitol did not enhance memory (data not shown). Other examples of metabolites showing highed levels in fecal samples from Lactobacillus inoculated mice include: galactonic acid (L. reuteri); D-xylose, glyceric acid, and methyl phosphate (L. plantarum); and uracil (L. brevis; Fig. S3A). Also, many of the GC–MS peaks corresponding to “carbohydrates”, presumably from the mouse chow, were lower in intensity in stool samples colonized with the Lactobacillus species suggesting that the components were being degraded by the inoculants (Fig. 3a).
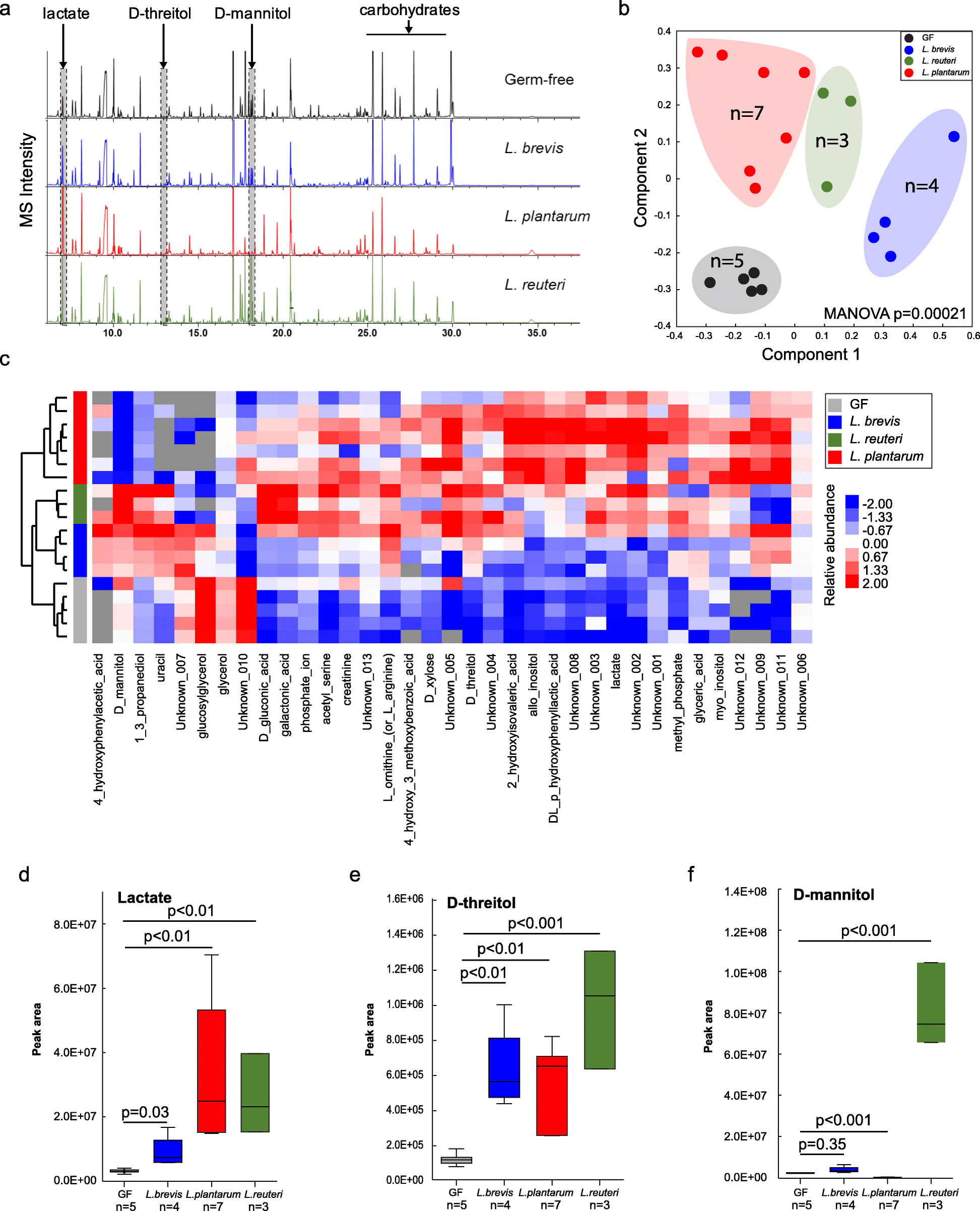
Fig. 3. Metabolomics analysis of fecal samples from Lactobacillus-colonized and germ-free mice.
a Representative GC–MS chromatograms of metabolite profiles in germ-free and Lactobacillus-colonized mouse fecal samples. Each chromatogram is a representative mass spectrometry profile from one cage of mice (4 mice/cage). b PCoA of metabolite profiles were measured in fecal samples. c Heatmap of metabolites differentiated between Lactobacillus-colonized and germ-free mice. d–f Relative abundance of select metabolites in fecal samples from individual mice. Error bars indicate mean ± SEM
To determine which metabolites could be possible mediators of the memory response, we also identified specific metabolites that were significantly higher in plasma and brain homogenates from the colonized mice compared to GF mice (Fig. S3). Fewer metabolite differences were found between species in plasma and brain, with the exception of higher plasma levels of arabitol, citric acid, glucose and L-tryptophan (L. plantarum; Fig. S3B). In the brain homogenates, species-specific differences included significantly higher levels of D-malic acid, dehydroascorbic acid, GABA, lactate, methyl phosphate, myo-inositol, and scyllo-inositol (L. plantarum); and glycine (L. reuteri; Fig. S3C). Examples of brain metabolites that were significantly higher in L. reuteri or L. plantarum inoculated mice compared to GF controls include glycerol, L-glutamic acid, L-serine, and N-acetyl-L-aspartic acid (Fig. S3C). Also, for both L. plantarum and L. brevis inoculated mice, 2,5-dihydroxypyrazine and citric acid were significantly higher in brain homogenates compared to GF controls (Fig. S3C).
Because we found an improved memory response with each of the three Lactobacillus species, we focused on fecal metabolites that were consistently higher across all three species compared to GF controls (Fig. S3), these included statistically (p < 0.05) higher levels of lactate (Fig. 3d), D-threitol (Fig. 3e), 2-hydoxyisovaleric acid, and acetyl-serine (Figure S3A). Interestingly, glycerol was significantly lower in fecal samples from mice inoculated with the three Lactobacillus species compared to GF controls (Fig. S3A). For the plasma samples, 1,5-anhydrohexitol, carbonate ion, pyruvic acid, and xylitol were significantly higher in all of the Lactobacillus inoculated mice compared to the GF controls (Fig. S3B). However, none of the identified metabolites were significantly higher in brain homogenates of Lactobacillus inoculated mice when compared to GF controls (Fig. S3C). Lactate did, however, have a trend towards higher levels in both plasma and brain, and was only significantly higher than GF controls (p < 0.05) in the brain samples from L. plantarum inoculated mice (Fig. S3C).
Based on these metabolite data and other studies of the role of lactate in memory formation [23], we hypothesized that lactate could be a mediator of the improved memory response in our trials because lactate is commonly produced by all lactobacilli. Also, lactate was recently shown to be higher in brain samples from rats that consumed a dietary supplement containing Lactobacillus and Bifidobacterium strains, and the rats had an improved memory as a result [8]. Therefore, we conducted an experiment to determine whether mice treated with dietary lactate had improved memory. CC042 mice, which have a relatively poor memory in the passive avoidance memory test (Fig. 1a), were treated with lactate through drinking water (0.5 g lactate/100 ml water) for 5 weeks. We found that dietary lactate treatment improved the average retention latency period of the CC042 mice from 92 s to 210 s (p = 0.01; Fig. 4).
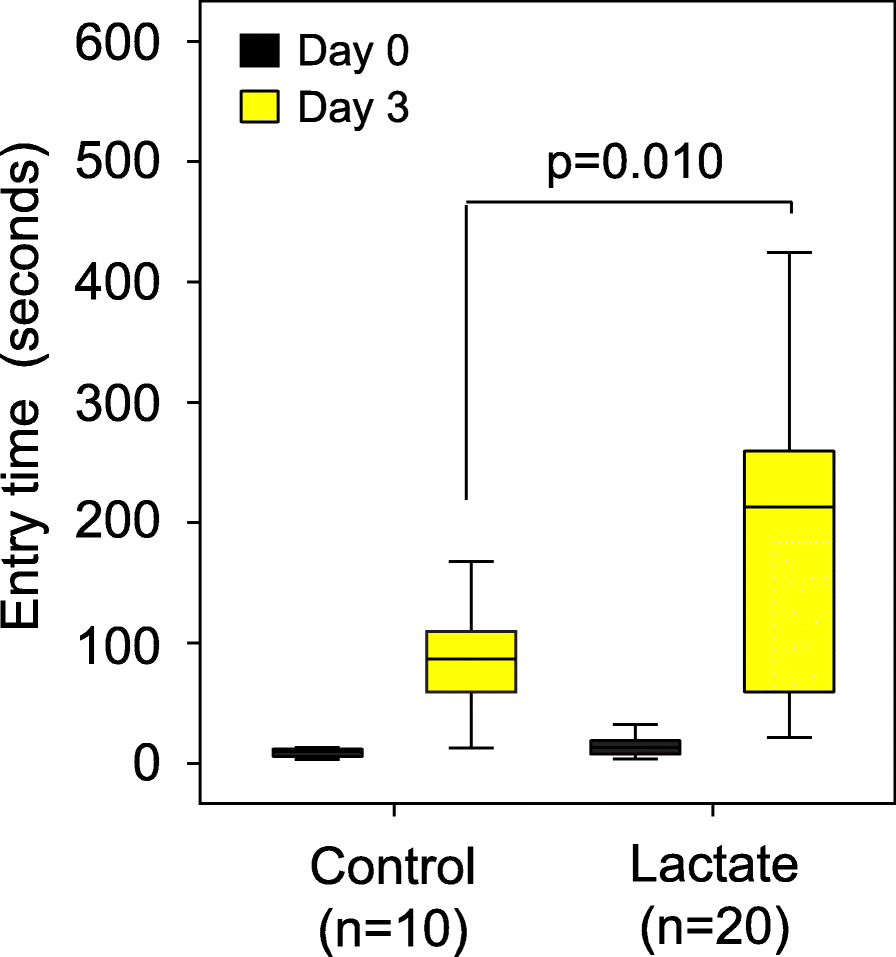
Figure 4. Dietary lactate treatment alone significantly boosted memory in CC042 mice.
C042 mice were treated with lactate through drinking water (n = 20; 0.5 g lactate/100 ml water) for 5 weeks or control (n = 20). Memory was assessed using passive avoidance. Dietary lactate treatment significantly improved the average retention latency period of the CC042 mice from 92 s to 210 s. Error bars indicate mean ± SEM
To further define the mechanism(s) underlying the improved memory response in mice following supplementation with Lactobacillus spp. or lactate, we quantified levels of GABA in the hippocampus proper, including the four Cornu Ammonis (CA) regions and the dentate gyrus (DG), using brain coronal sections taken from the treated mice. We chose to focus on the hippocampus as it is an essential area for the acquisition and formation of new memories, where GABA, the main inhibitory neurotransmitter in the brain, plays a critical role [24, 25]. As mentioned above, using metabolomic analysis we found that GABA levels were significantly higher in brain homogenates of mice inoculated with L. plantarum, but not with the other two species; although at a lower significance threshold GABA levels were also higher in the L. reuteri samples (p = 0.065; Fig. S3C). Using immunofluorescence, we quantified GABA expression in the hippocampus (Fig. S4) and GABA expression was compared between the GF controls and the three Lactobacillus-colonized mice groups (Fig. 5a, Fig. S4). All of the Lactobacillus-colonized mice showed higher fractions of cell bodies that expressed GABA in the hippocampus, compared to the germ-free control mice (Fig. 5b). However, our metabolite data showed no significant increases in GABA in stool or plasma samples of the inoculated mice compared to controls. Interestingly, we found that lactate treatment alone also increased the fractions of cell bodies that expressed GABA compared with control mice from 32% to 43% (p = 0.042; Fig. 5c), supporting the hypothesis that lactate could serve as the metabolic conduit between the gut and the brain.
.../...
.











































