.
O P E N A C C E S S S O U R C E : Aging Research News
Highlights
• Ageing is a highly complex process driven by a multitude of factors.
• Advanced methods are emerging allowing us greater understanding of ageing.
• These include novel model organisms, high-throughput experimental approaches and machine learning for analysis of large and complex data.
• Given the intricate nature of ageing and the increase in data availability, these methods will become increasingly important in the coming years.
Abstract
Ageing is arguably the most complex phenotype that occurs in humans. To understand and treat ageing as well as associated diseases, highly specialised technologies are emerging that reveal critical insight into the underlying mechanisms and provide new hope for previously untreated diseases. Herein, we describe the latest developments in cutting edge technologies applied across the field of ageing research. We cover emerging model organisms, high-throughput methodologies and machine-driven approaches. In all, this review will give you a glimpse of what will be pushing the field onwards and upwards.
1. Introduction
Ageing is the most profound risk factor for most diseases, and methodologies to study the ageing process are therefore of critical importance. In the last one hundred years the field has experienced rapid progress from the discovery that dietary interventions alter the pace of ageing in rats to the development of artificial intelligence algorithms that can predict age with high accuracy (Bobrov et al., 2018; McCay and Crowell, 1934). However, the field has branched out immensely with numerous sub-specialties focusing on a variety of themes within ageing research all with specialised techniques. Here we will discuss some of the most cutting-edge technologies in three broad areas: emerging model organisms, high-throughput methodologies for organismal investigations and machine learning approaches.
2. Model systems for ageing research
Research on ageing has always been driven by studies on a wide variety of different model systems (Table 1). These studies have led to the identification of drivers of the ageing process as well as to the identification of novel ageing interventions. Each model system provides its unique strengths, however in particular, cross-species comparative studies have helped to further understand evolutionary conserved ageing mechanisms. Rodent animal models have been a cornerstone in ageing research but have been extensively reviewed by others (Folgueras et al., 2018; Kõks et al., 2016; Mitchell et al., 2015) and will not be further discussed here.
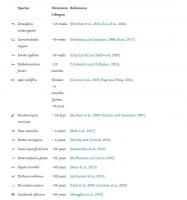

Table 1. Examples of organisms and human disease models in ageing research. In red are understudied organisms that could give unique insight into ageing. Maximum lifespans for animal species extracted from AnAge database (Tacutu et al., 2018). Human disease models are more variable in nature and hence mean and/or median lifespan is included for these based on publications mentioned in table.
2.1. Naturally short-lived ageing-model organisms
Traditional model organisms such as Drosophila melanogaster and Caenorhabditis elegans have contributed tremendously to our current knowledge about the biology of ageing. The advantages of such organisms are plentiful, as they are cheap, simple to handle and easy to genetically manipulate. C. elegans straight forward germline genetics as a self-fertilizing hermaphrodite is well-described, the advantages and disadvantages of this model organism for ageing studies has been much discussed (Johnson, 2003), however the ability to easily establish RNAi libraries is a key advantage of this organism (Timmons et al., 2001). D. melanogaster has likewise been widely used in ageing research and while being a short-lived organism has physiology that more closely resemble mammals than nematodes, including for example replicating cells (Helfand and Rogina, 2003).
A testimony to the importance of short lived organisms was the central discovery that a mutation in a single gene, Age-1, results in lifespan extension in C. elegans (Friedman and Johnson, 1988). This was followed by the identification of multiple genes and compounds that influence health- and lifespan in D. melanogaster and C. elegans (Hall et al., 2019; Uno and Nishida, 2016). The ease of comparing different age groups makes short-lived animals of further interest for ageing studies, as highlighted in recent studies that revealed a correlation between the microbiota composition and the health- and lifespan in D. melanogaster and C.elegans (Clark et al., 2015; Han et al., 2017). Further, single-cell transcriptomic changes were studied in the ageing fly brain which revealed decreased gene expression as well as changes in cell type composition with age (Davie et al., 2018). Clearly, these short-lived organisms remain a strong model system for the mechanistic understanding of ageing.
Besides these traditional models other less studied short-lived animals have recently been recognised for providing insight into the ageing phenomenon. For instance, the African turquoise killifish has proven to be a unique platform for studying ageing mechanisms (Harel et al., 2015). Notably, the lifespan of different turquoise killifish strains varies even under identical conditions (4–8 months) and has been shown to be influenced by genes that are linked to the sex-determining region (Valenzano et al., 2015). Moreover, middle-aged fish treated with the gut microbiota of young fish showed improved lifespan as well as increased locomotor activity, which was validated by video-tracking approaches (Smith et al., 2017).
Another interesting organism that provides unique opportunities to study ageing mechanisms is the honey bee Apis mellifera. A. mellifera has a quite variable lifespan, which is dependent on seasonal aspects and on their caste and task within the colony most strikingly with a ∼10-fold increase in lifespan when a female larvae, that is set to become a queen, is exclusively fed a diet of royal jelly (Münch and Amdam, 2010). Bees provide opportunities to also study social behavioral effects on the ageing process, as for instance, older bees repositioned to nursing tasks revealed a reversal of some brain ageing features (Baker et al., 2012). The possibility to track short-lived animals over their whole lifespan as well as over several generations makes them particularly interesting for ageing studies.
Clearly genes play an important role in impacting the lifespan of organisms. It is therefore critically important to validate pathways found in simpler organisms with changes in mammals and preferably humans. Nevertheless, these short-lived model organisms have proven potent to delineate mechanisms and pathways central to human ageing. One small example is the importance of the insulin receptor and nutrient sensing in mammalian longevity that were catalyzed by studies on daf-2 mutants in C.elegans (Kenyon et al., 1993) and InR mutants in drosophila (Tatar et al., 2001).
2.2. Naturally long-lived ageing model organisms
Another group of organisms that present an exciting opportunity to study basic molecular mechanisms of ageing and the pathomechanism of age-related diseases are naturally long-lived animals. Since some long-lived animals tend to show a decreased rate of age-associated diseases such as cancer, it suggests that they may evolved specific protection mechanisms against stress. The lack of correlation between cancer risk and body size or lifespan is known as Peto’s paradox (Peto et al., 1975). Genomic studies on the African elephant have tried to explain this phenomena through the identification of gene duplication events resulting in 20 copies of the tumor suppressor gene TP53, whereas the human genome contains one copy (Abegglen et al., 2015; Sulak et al., 2016). The expression of multiple TP53 genes leads to enhanced p53 signaling, revealed by increased apoptosis signaling upon DNA damage induction in elephant cells. Increased genome maintenance capacity has been also revealed for other long-lived species such as the bowhead whale (Keane et al., 2015), bats (Zhang et al., 2013) and naked mole rats (MacRae et al., 2015). Importantly, not all long or short-lived species have had their genomes sequenced (for example S. microcephalus), and hence there is undoubtedly vast discoveries hidden away in the genomes of these organisms.
However, most of these long-lived animals are challenging for ageing research due to the difficulty in keeping them in captivity and the high costs to implement experiments. Nevertheless, latest advances in sequencing technologies have allowed the expansion of cross-species comparative studies and investigations on so far un-investigated long-lived model organisms (Seim et al., 2014; Tian et al., 2019). Interestingly, there are still unexplored animals with extreme longevity such as several species of antarctic seasponge (such as Anoxycalyx (Scolymastra) joubini) that may live for several thousand years (Dayton, 1979; Gatti, 2002). However, a creature dwelling at the bottom of the Antarctic ocean has a vastly different environment than humans which will impact ageing in numerous ways. For this reason, investigating animal models living in habitats very similar to humans may be of interest. Here, the recently commenced project looking into interventions targeting ageing in companion dogs may be of particular importance (Kaeberlein et al., 2016).
2.3. Genetic components of human ageing
The study of inherited human premature ageing disorders has emerged as a seminal approach in ageing research (Kipling et al., 2004). Importantly, the identification of the underlying genetic mutations revealed that these disorders are characterised by compromised genome integrity, corroborating the idea that accumulation of DNA damage possesses a key role in ageing (Hoeijmakers, 2009). Notably, these are monogenic diseases, thus, paving the way for the illumination of the specific molecular defects involved in ageing and further allowing for direct manipulation of the implicated pathways. Common approaches such as utilizing fibroblasts or stem cells derived from patients, have yielded critical information into the disease mechanisms as well as normal human ageing processes. For instance, the cellular phenotype of Hutchinson-Gilford progeria syndrome, as well as Cockayne Syndrome, has been largely elucidated (Cleaver, 1969; Merideth et al., 2008; Scaffidi and Misteli, 2005; Scheibye-Knudsen et al., 2012). In addition, some mouse models of premature ageing have further underscored the role of declining levels of the metabolite NAD+ and NAD+-dependent proteins in age-related metabolic dysfunction (Fang et al., 2014; Scheibye-Knudsen et al., 2014). Importantly, age-dependent NAD+ decline, as well as a number of other features observed in premature ageing models, are also observed in normal ageing indicating that these may represent suitable models of human ageing (Gomes et al., 2013; Mouchiroud et al., 2013).
Interestingly, mouse models of premature ageing do not recapitulate all phenotypic features seen in the human diseases and often display milder phenotypes indicative of more redundancy in ageing-associated pathways in mice. In addition, age-related neuropathologies are currently poorly reflected in mouse models (Burns et al., 2015; Jucker, 2010).
An alternative to premature ageing diseases, are studies with exceptionally long-lived people. Centenarians have long been a subject of curiosity, owing to the mystery of how these people retain their health at very advanced age. Here, research has identified some of the factors, genetic as well as environmental, which appear to protect from age-related disease. Of importance, studying centenarians permits the consideration of key variables, including demography, population genetics, lifestyle, and cultural habits, for instance highlighted by findings illustrating health-promoting effects of social relationships and higher socioeconomic status (Seeman and Crimmins, 2001; Yashin et al., 1999). Accordingly, the study of centenarians has shed novel light on the role of several molecular mechanisms in ageing, including immunosenescence (Effros et al., 1994), inflammation (Franceschi et al., 2005), gut microbiota (Biagi et al., 2010), and mitochondrial DNA genetics (Salvioli et al., 2008). There is an apparent familial trait for extreme longevity (Perls et al., 2000), and close to 300 single nucleotide polymorphisms (SNPs) have been implicated (Sebastiani et al., 2013). In particular, SNPs in the genes of apolipoprotein E (APOE) and the forkhead box O3A (FOXO3A) have been extensively studied and found to be significantly associated with exceptional longevity (Revelas et al., 2018). However, the significant variants display relatively modest effects sizes, supporting the idea that the genetic component consists of a large number of genetic modifiers, each contributing with a minor effect on human ageing (Sebastiani and Perls, 2012). Accordingly, the phenotype of centenarians is highly complex, and the molecular core remains poorly understood. The emergence of new high-dimensionality and ‘Omics’ technologies, including genetics, epigenetics, metagenomics, metabolomics, proteomics, glycomics, etc. (Lorusso et al., 2018), has allowed for the identification of genetic signatures that characterise the nature of centenarians. For instance, lipidomic profiling of individuals with exceptional longevity using mass spectrometry has been able to discriminate between adult, aged and centenarian with a 90 %–100 % accuracy (Jové et al., 2017; Pradas et al., 2019).
In summary, animal and human models of extreme ageing have given us instrumental insight into ageing and will unquestionably continue to help us to deepen our knowledge of the ageing process.
3. Machine driven approaches to the Ageing riddle
Ageing represents the most complex combination of molecular, cellular and organismal features seen in organisms (Andreassen et al., 2019). Given the vast complexity of the ageing process machine learning algorithms are emerging as key tools for prediction, discovery and treatment investigations. For instance, both of the first algorithms to accurately determine the age based on epigenetic changes, the Horvath and Hannum clocks, used elastic net regression (Hannum et al., 2013; Horvath, 2013). Notably, using deep neural networks combined with feature importance analysis it is possible to determine which features contribute most to the predictive power of an algorithm, a technique that revealed that albumin is a strong predictor of ageing when considering common blood samples values (Putin et al., 2016). While blood samples are readily available, age-prediction from even simpler datasets such as facial photographs appear to be able to predict ageing with high accuracy (Bobrov et al., 2018). In the context of treatments, machine learning is showing great potential in terms of drug design (Zhavoronkov et al., 2019), target identification (Madhukar et al., 2019) and outcome prediction (Chekroud et al., 2016). Thus, in the last decades numerous great advances has been made in machine driven approaches to ageing (See Fig. 1 for a timeline).

Fig. 1. A timeline of recent methodological advances in ageing research.
To facilitate understanding of the plenitude of organisms and their different ageing phenotypes, there is a great need to observe ageing models utilizing longitudinal tracking to accurately capture the complex set of parameters required for the characterization of ageing across different species. Here, we describe state-of the art methods meeting this demand in different fields, from cell-based and microscopy assays, microfluidics, automatic analyses of animal models with computer vision technologies, and the assessment of human longevity in clinical studies (Fig. 2).

Fig. 2. New technologies for monitoring age- and associated features. Automation and high throughput methodologies are being deployed across the ageing research space: from single cell approaches to human investigations.
3.1. Monitoring ageing using microscopy-based assays
Robotics and automation are increasingly being used in biological investigations likely allowing higher reproducibility and the possibility of casting a much wider net for mechanistic exploration. A prime example is whole genome CRISPR or siRNA screens where >20,000 genes are manipulated, and readouts are typically made using high-content microscopy. High-content microscopy can also be used in more specialised approaches such as the Comet-chip assay were DNA damage can be measured in individual cells embedded in microwells (Albert et al., 2016; Sykora et al., 2018; Wood et al., 2010). High-content microscopy has also been applied to drugs screens for compounds that reduce ageing features such as beta-galactosidase expression, cellular features of progeria or neurite outgrowth in iPSC-derived neurons (Kubben et al., 2016; Sherman and Bang, 2018; Vatolin et al., 2019). These types of investigations are increasingly being adapted to more complex cell-based assays such as in 3D organoid cultures and even pursued in high-throughput ‘organ-on-a-chip’ applications (Probst et al., 2018; Williamson et al., 2018). The use of these assays is thus quickly becoming essential in ageing research, particularly in the area of interventions testing.
The short life cycle of single-cell organisms makes them a particularly interesting model for ageing research. Despite their inherent simplicity, monitoring a panel of different features of unicellular organisms during long-term culture can be challenging and labor-intensive. Since the early attempts of tracing the ageing process in yeast in the late 1950s (Mortimer and Johnston, 1959), time-consuming yeast replicative lifespan assays (taking up to 4 weeks) have been simplified through extensive automation. The most promising approach to date combines microfluidic platforms with continuous high-resolution imaging (Lee et al., 2012; Zhang et al., 2012). By developing the automated tracking of entire life cycles of single yeast cells in microfluidic systems such as the high-throughput yeast ageing analysis chip (HYAA-Chip) (Jo et al., 2015) or the more advanced Yeast Replicator (Liu et al., 2015), new prospects have opened up in phenotype tracing. By allowing the simultaneous life-long monitoring of up to 16 different strains of yeast, automated microfluidic platforms overcome former limitations in the usage of yeast as broad genetic screening platforms in ageing research. Going beyond the scope of observing longevity-related changes in morphology, microfluidic techniques also allow for the simultaneous monitoring of gene expression patterns over lifespan (Kaiser et al., 2018). Moreover, they can help to estimate the relevance of different parameters within the ageing process, including different growth environments or cell-to-cell heterogeneity, among others, depending on the set-up of the respective microfluidic platform (Chen et al., 2017).
Microfluidic culture systems are not only attractive for unicellular organisms, but hold great potential for monitoring nematodes and potentially other species. Foremost, culturing of C. elegans in microfluidic systems has set an example for the great advantages of automated long-term culture (Atakan et al., 2019). The novel microfluidic culture platform allows for automated video tracking over entire lifespans, as well as for individual phenotyping and simultaneous implementation of drug treatments at a high-throughput level. In this manner, tracking of and interference in the ageing process can be achieved at the same time.
3.2. Non-invasive, automated longitudinal tracking in model organisms of ageing
A battery of methods exists to assess model organisms over long time periods; however, their application could be implemented to even greater potential. This recent realization and progress is exemplified by a comprehensive overview of age-related readouts in mice (Bellantuono et al., 2020) D. melanogaster (Gaitanidis et al., 2019), and C. elegans (Stroustrup et al., 2013). These readouts can be expanded on in various forms: continuous deep-learned computer vision tracking of model organisms with age (www.tracked.bio), comprehensive in vivo imaging and the combination of these systems with a spectrum of physiological sensors to rapidly modulate ageing.
An approach that has been repeatedly verified across various species is computer vision technology. This technology has already been widely applied to various model organisms (Nakamura et al., 2015; Robie et al., 2017), however, its application to the longitudinal measurement of ageing is lacking, albeit there are several applications on the rise (Stroustrup et al., 2013). The inspiration for such long-term tracking comes from knowledge gained from piecemeal and cross-sectional applications from, for instance, animal behavior experiments and human gait analysis (Bair et al., 2019; Studenski et al., 2011). The same technology has been applied longitudinally on a wild population of chimpanzees (Schofield et al., 2019). Hence, applying longitudinal tracking for lab models represents a great opportunity to further our understanding of ageing and particularly towards the development of interventions; the last piece to this puzzle is automated methods.
Another mode to longitudinally assess model organisms is via in vivo imaging, such as the use of MRI, microCT, and other in vivo imaging methods (Dall’Ara et al., 2016). The techniques themselves are not new and have been extensively used in mammalian animal models, however, progress in MRI techniques now allow visualization and quantification at a single cell level (Chung et al., 2020; Tsurugizawa et al., 2020). Given the non-invasive nature of these techniques, they prove advantageous to other strategies to obtain molecular and physiological dynamics i.e. purely histological studies, resorting to traditional biochemical techniques, etc. In this context, non- or minimally-invasive sensors measuring a variety of outputs are also being employed from telemetry ECG/EEG (Axsom et al., 2019), activity/temperature monitors (Meyer et al., 2017), to in vivo metabolism (Brockway et al., 2015; Pedersen et al., 2018). Altogether, these physiological sensors can be used for extracting much richer information from each of the above referenced models.
3.3. Advances in comprehensive health assessment in humans
With the rapid increase in elderly people across the globe it is a priority to develop automated ways to assess quality of life for improving life- and healthspan within longitudinal human studies. One example of health function acquisition and assessment among elderly people is through a pipeline of Comprehensive geriatric assessment (CGA) that includes measurements of multiple parameters that take into account both mental and physical states of the studied population (Ellis et al., 2017). Although this approach aims to identify the most effective ways to improve quality of life and independence among elderly populations worldwide, it strongly depends on the country where the assessment is made as well as on their healthcare system (Wilhelmson et al., 2020). Moreover, accurate assessment of multiple parameters can be time- and cost-intensive. Therefore, “every day” tracking systems as well as more precise and automated approaches for large-scale data analysis are needed to more effectively evaluate health function both of individuals and of populations.
An example of a comprehensive and multidimensional longitudinal study to understand ageing processes in humans is the Baltimore longitudinal study of Ageing (BLSA). This program considers biological, behavioral and environmental factors that affect changes that occur during normal ageing. The advantage is that BLSA is a continuous study on volunteers that includes research on different aspects of ageing and age-associated diseases (Lin et al., 2011; Nastasi et al., 2018; Vidoni et al., 2018). Another example of a comprehensive study is via personalised healthcare monitoring using different wearable devices that may track diurnal rhythm patterns, body temperature, heart rate, etc. These devices can be used not only for daily monitoring of standard health quality parameters, but also for preventing unexpected health failure, for instance for people with epilepsy (Ryvlin et al., 2018). Moreover, wearable devices that measure heart rate show quite good correlation with standard ECG (Georgiou et al., 2018).
The direction of comprehensive health assessment and personalised medicine is also actively developed by industry in the application of multi-omics approaches combined with machine learning to develop novel tools for accurate assessment of personal health (Hou et al., 2020; Shomorony et al., 2020). This approach also includes coupling whole genome sequencing with full-body MRI using advanced imaging protocols and data quantification to find age-related chronic disease risk factors (Perkins et al., 2018). The development of AI-based whole-body MRI has also progressed rapidly in other age-related diseases like cancer (Lavdas et al., 2019).
In summary, the combination of high-throughput and automated methodologies can create a finer and more accurate understanding of the progression of ageing on the single-cell, whole model organism, and human level. Furthermore, the development of interventions to alleviate ageing phenotypes may be identified faster and more cheaply with such technologies.
4. Machine learning for biomarker discovery
One of the key roadblocks in the development of ageing research therapeutics is the discovery of robust biomarkers. Machine learning techniques as well as novel image- and text-based mining strategies are emerging to assist in biomarker discovery in the ageing field (Fig. 3).
Fig. 3. Machine learning in ageing research. Text-mining and image-based data repositories are increasingly being used in combination with AI-assisted and machine-learning techniques to support the search for novel biomarkers that characterise ageing. Such age-predictive clocks based on a multitude of these data inputs have the power to delineate both pathogenic and delayed ageing, and hold promise to stratify patients and/or test ageing interventions.

















































