.
O P E N A C C E S S S O U R C E : nature
Abstract
Mindfulness and meditation techniques have proven successful for the reduction of stress and improvement in general health. In addition, meditation is linked to longevity and longer telomere length, a proposed biomarker of human aging. Interestingly, DNA methylation changes have been described at specific subtelomeric regions in long-term meditators compared to controls. However, the molecular basis underlying these beneficial effects of meditation on human health still remains unclear. Here we show that DNA methylation levels, measured by the Infinium HumanMethylation450 BeadChip (Illumina) array, at specific subtelomeric regions containing GPR31 and SERPINB9 genes were associated with telomere length in long-term meditators with a strong statistical trend when correcting for multiple testing. Notably, age showed no association with telomere length in the group of long-term meditators. These results may suggest that long-term meditation could be related to epigenetic mechanisms, in particular gene-specific DNA methylation changes at distinct subtelomeric regions.
Introduction
In recent years, the use of mindfulness and meditation techniques have been increasing in western societies with the aim of reducing stress and improving overall health1,2,3,4. Mindfulness-based interventions have proved their beneficial effect on a number of medical and psychological conditions, including depression, anxiety, and immune disorders5,6,7,8. In addition, it has been proposed that meditation techniques could positively affect longevity9,10. In fact, intensive meditation training has been associated with an increase in telomerase activity11 and longer telomere length in blood cells12,13,14,15,16, which is considered a candidate biomarker of human aging.
Telomeres are DNA-protein complexes that protect the end of linear chromosomes from degradation, fusion, or DNA repair processes17,18,19. Telomeres shorten with every somatic cell division because of the end replication problem19,20, but this erosion is partially compensated by the action of telomerase, an enzyme complex that adds TTAGGG hexanucleotide repeats to the telomeric DNA17,21. However, telomerase expression levels in mammalian adult cells are not sufficient to preserve the original length of telomeres, resulting in the progressive shortening of chromosomes throughout life20,22,23,24. Telomere shortening has been associated with age-related conditions25,26,27,28, and telomere length is hypothesized to be a biomarker of aging and age-related morbidity29,30,31,32,33,34.
In line with other reports, our group previously observed a positive relationship between meditation practice and longer telomere length in peripheral blood cells13. Moreover, in a previous work to gain insight into the molecular mechanisms of meditation, we profiled genome-wide DNA methylation changes in long-term meditators and identified a set of 64 differentially methylated regions (DMRs), corresponding to 43 genes, when compared to controls35. Almost half of the DMRs (48.4%) were directly linked to common human diseases, including neurodegenerative disorders such as Alzheimer’s and Parkinson’s diseases. Surprisingly, up to 23.4% of these DMRs were found to be located at subtelomeric regions (hypergeometric test fold-enrichment = 1.3; p-value < 0.05). However, the importance of these epigenetic changes within the subtelomeric regions in meditators remains unclear.
Increasing evidence supports the idea that epigenetic changes in subtelomeric regions may be linked to telomere length. For instance, in blood cells from healthy middle-aged men it was found that CpG sites whose methylation level was associated with telomere length were significantly enriched at subtelomeric regions36. Most importantly, TET (Ten-eleven translocation) enzymes which are responsible of converting 5-methylcytosine to 5-hydroxymethylcytosine, have been proved crucial in telomere maintenance by modulating subtelomeric methylation levels37. Additional evidence on the potential role of DNA methylation in telomere homeostasis shows that mouse embryonic stem cells lacking DNA methyltransferases have decreased global methylation in subtelomeric regions and dramatically elongated telomeres38. On the contrary, telomere shortening to a critically short length leads to epigenetic defects at mammalian telomeres and subtelomeres, such as decreased subtelomeric DNA methylation39,40.
A number of pathological conditions support the possible involvement of subtelomeric DNA methylation in telomere length stability. Human mutations on the DNA methyltransferase DNMT3B [OMIM, * 602900], which cause immunodeficiency-centromeric instability-facial anomalies syndrome 1, are associated with subtelomeric global hypomethylation along with accelerated telomere shortening of fibroblasts41,42. Both direct and inverse correlation has been described for various types of human cancer between subtelomeric DNA methylation levels and telomere length43,44. To sum up, DNA methylation levels at subtelomeric regions might play a role in maintaining telomere length, among other mechanisms. However, the potential effect that subtelomeric DNA methylation levels may exert on telomere length and the underlying mechanisms remain unknown.
The aim of the present study was to better understand the relevance of subtelomeric DNA methylation changes in long-term meditators. To that end, we tested the association of telomere length with DNA methylation levels and age in long-term meditators and controls for this set of DMRs.
Results
Subjects characteristics
The general characteristics of subjects included in the study are shown in Table 1. Socio-demographic characteristics including age, gender, ethnicity, and BMI were equally distributed between groups. However, as expected, there were significant differences in the amount of practical experience of meditation and also in all the psychological health-related variables referred to below (see Methods section).

Table 1. Characteristics of study participants: Figures represent means, standard deviations (†), and the p-value associated with a t contrast between the control group and the meditators group, except for sex and education, where the figures represent frequencies and percentages (‡) and the p-value associated with a χ2 contrast. Range indicates the bounds of the scales for each psychological variable.
Relationship between DNA methylation levels and telomere length
From our genome-wide DNA methylation differential analysis between long-term MMs and controls35, we selected the group of 14 DMRs located in subtelomeric regions, involving chromosomes 2, 4, 5, 6, 7, 8, 16, 19, and 20. We define subtelomeric regions as those regions at either end of each chromosome with a length of 3000 kb. We selected this subtelomeric length as an operational definition in order to perform the bioinformatics analysis, since the exact bounderies for subtelomeres have not yet been well established. This group represents 23.4% of the total DMRs (hypergeometric test fold-enrichment = 1.3; p-value < 0.05) that were identified in long-term MMs compared to controls. Genes and DNA methylation levels for these regions are shown in Table 2.
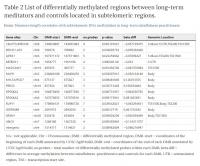
Table 2. List of differentially methylated regions between long-term meditators and controls located in subtelomeric regions: NA = not applicable, Chr = Chromosome, DMR = differentially methylated region, DMR-start = coordinates of the beginning of each DMR annotated by UCSC hg19 build, DMR-end = coordinates of the end of each DMR annotated by UCSC hg19 build, no.probes = total number of differentially methylated probes within each DMR, beta.diff = difference in average methylation between mindfulness-practitioners and controls for each DMR, UTR = untranslated region, TSS = transcription start site.
The raw differences between groups showed that telomere length was longer in meditators (Mn = 10.47; SD = 0.86) compared to controls (Mn = 9.87; SD = 0.94), without any statistical significance, but showing a strong trend (t = −1.94; df = 32; p = 0.061), with a moderately high ES (d = 0.67). However, the differences appeared to be significant (t = −3.44; df = 32; p = 0.002) after controlling for age, with a very high ES (d = 1.25). We then asked whether telomere length was correlated with gene-specific DNA methylation levels. In the group of long-term MMs, we found a positive raw correlation between telomere length and DNA methylation levels at the GPR31 gene (r = 0.58, p = 0.014), whereas an inverse correlation was shown between telomere length and DNA methylation levels at two distinct loci: SERPINB9 gene (r = −0.64, p = 0.006) and an intergenic CpG island within the subtelomeric region of chromosome 4 short arm (chr4:1514317–1514621, GRCh37/hg19 assembly) (r = −0.51, p = 0.036) (Fig. 1). After correction for age, the significant correlations remained (Table 3). To test for multiple comparisons, we draw at random 1000 sub-samples of 14 not differentially methylated regions within subtelomeric regions. The proportion of such random sets showing 3 or more hits with absolute r value greater than or equal to 0.51 (the minimum value for our selection) was 0.052 (>0.05). Therefore, the possibility of a false positive result cannot be completely excluded.

Figure 1. Relationships between intergenic (chr4: 1514317–1514621), GPR31, and SERPINB9 DNA methylation levels and telomere length, according to group: The intergenic region (chr4: 1514317–1514621) is in the first row, GPR31 in the second, and SERPINB9 in the third. Long-term meditators are in the first column and controls are in the second. Methylation levels are represented in the horizontal axis (X), and telomeres length in the vertical axis (Y).
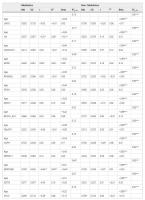
Table 3. Explanatory power of gene methylation on the length of long telomeres according to group: Md = mean. SD = standard deviation. r = Pearson’s raw correlation between telomere length and gene methylation. R2 = bivariate determination coefficient (dependent variable: telomere length; independent variable: genes methylation). R2y.12 = multiple determination coefficient (dependent variable: telomere length; independent variables: age and gene methylation). Beta = standardized slope when using multiple regression models. *p < 0.05. **p < 0.01. ***p < 0.001.















































