.
O P E N A C C E S S S O U R C E : Neuroscience Insights
Abstract
The number of age predictors based on DNA methylation (DNAm) profile is rising due to their potential in predicting healthspan and application in age-related illnesses, such as neurodegenerative diseases. The cumulative assessment of DNAm levels at age-related CpGs (DNAm clock) may reflect biological aging. Such DNAm clocks have been developed using various training models and could mirror different aspects of disease/aging mechanisms. Hence, evaluating several DNAm clocks together may be the most effective strategy in capturing the complexity of the aging process. However, various confounders may influence the outcome of these age predictors, including genetic and environmental factors, as well as technical differences in the selected DNAm arrays. These factors should be taken into consideration when interpreting DNAm clock predictions. In the current review, we discuss 15 reported DNAm clocks with consideration for their utility in investigating neurodegenerative diseases and suggest research directions towards developing a more optimal measure for biological aging.
Introduction
In contrast to the steady pace of chronological age, the pace of biological age varies among individuals and may predict distinct aspects of aging at different life stages. For example, biological aging later in life reflects the link with disease, morbidity, and mortality, whereas in youth/midlife, it might predict healthspan or cognitive/physical decline (eg, individuals aging more rapidly are less cognitively/physically able).1,2 As chronological age does not sufficiently represent fundamental aging processes, methods to measure biological aging have been developed, which is important for assessing strategies to slow down biological aging and extend healthspan. A reliable measure of biological age could be a factor in predicting disease onset in presymptomatic carriers of causal mutations, and assist in the development of preventive rather than therapeutic strategies.1
Technical breakthroughs have led to the discovery of several molecular markers of aging, including epigenetic biomarkers.3-6 For instance, genome-wide RNA-interference-based screening in Caenorhabditis elegans revealed a conserved epigenetic mechanism, which implicated 59 genes as modulators of age-related behavioural deterioration rate.7 Two neuronal genes were among the most prominent hits: epigenetic reader (BAZ-2) and histone 3 lysine 9 methyltransferase (SET-6), which could accelerate behavioural deterioration by repressing the expression of nuclear-encoded mitochondrial proteins. Importantly, the expression of human orthologues (BAZ2B and EHMT1) in the frontal cortex increases with age and correlates with Alzheimer disease (AD) progression.
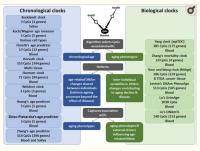
Among biomarkers of aging, such as telomere length (TL), metabolomic, transcriptomic and proteomic variations, the most promising are based on the DNA methylation (DNAm) of cytosines at CpG dinucleotides,8 representing one of the key epigenetic mechanisms altering gene expression or splicing. The cumulative assessment of DNAm levels at age-related CpGs could be used as a DNAm clock,6,9-13 which may mirror biological aging (Figure 1). Although some clinical biomarkers outperform DNAm clocks in reflecting morbidity and mortality,14 the advantage of DNAm clocks is their ability to measure either multitissue or cell-/tissue-specific aging. DNAm clocks could help explain why some individuals stay healthy, whereas others develop age-related neurodegenerative diseases.
Figure 1. Comparison of chronological vs biological DNAm clocks. Each DNAm clock is developed using a unique training model, including a variable number of CpGs, tissue source of DNA and corresponding age-related measures. While chronological DNAm clocks reflect age-related DNAm changes that are shared between individuals and are expected to reflect the intrinsic aging process, biological DNAm clocks reflect age-related DNAm changes that vary between individuals and are expected to capture associations with specific age-related phenotypes and external drivers that may influence age-related DNAm.
Several studies support the link between DNAm clocks and biological age. DNAm-age acceleration (difference between DNAm-age and chronological age) was associated with major neurodegenerative diseases, such as AD,15 Parkinson disease (PD),16 Huntington disease17 and amyotrophic lateral sclerosis (ALS).18,19 Similarly, HIV-infected individuals exhibit premature aging based on methylome-wide changes and Horvath DNAm clock.20,21 Furthermore, individuals with Werner or Down syndromes also display accelerated DNAm clocks.22,23 In contrast, DNAm-age in centenarians is on average 9 years younger than their chronological age.24
However, it is mostly unclear what the underlying molecular mechanisms of DNAm clocks are. Do they reflect similar aspects of the aging process? What is their capacity to predict risk of decline before disease onset and therapeutic effectiveness aiming to extend healthspan? Here, we provide an overview of DNAm clocks, including their application in neurodegenerative diseases, potential confounders and gaps in the current knowledge.
Comparison of Different DNAm Clocks
Most DNAm clocks are based on a limited number of CpGs, which are widely distributed throughout the genome and mainly selected from the 27K or 450K BeadChips that have currently been replaced by the more informative EPIC chip covering 850K CpGs (Table 1). Age-related CpGs are overrepresented in the proximity of Polycomb-binding regions and promoters, which are key regulators of gene expression.6,11,13 The little overlap in CpGs between different DNAm clocks25-27 might be the result of several factors (eg, cell-/tissue-specific differences) and explain why some clocks are capable of catching certain outcomes better than others. In addition, the applied arrays might not include the most informative age-related CpGs.6,14 Many DNAm clocks are obtained from linear regression algorithms trained against chronological age6; however, clocks too perfectly trained on chronological age would not contain information on interindividual variability in biological age.5,6 Finally, some DNAm clocks focus on the comparison between DNAm levels and physiological measures of biological age (eg, cholesterol levels), which may be the consequence of confounding factors (eg, obesity) rather than representing aging itself.28
Table 1. An overview of the different chronological and biological DNAm clocks, including the number of CpGs and genes used to build the clocks, the age range in which they can be applied, as well as the sample size, tissues and platforms used to develop the different DNAm clocks.
Chronological DNAm clocks
An overview of all 9 reported chronological DNAm clocks is presented in the Table 1. The first DNAm clock was built by Bocklandt et al,9 using saliva DNA. The study identified 88 CpGs that correlated with the chronological age of 34 male identical twins (age, 21-55), of which 3 were replicated in a general cohort (n = 60; age, 18-70).9 However, DNAm clocks composed of only a few CpGs cannot be reliably used for multiple tissues.5 For instance, a high average error of age prediction (11 years) was reported for the ‘epigenetic-aging-signature’ of Koch and Wagner,29 containing 19 CpGs. Notably, the DNAm-age estimator of Weidner et al13 consists of only 3 CpGs, the analysis of which by bisulphite pyrosequencing revealed age prediction with an average error of 4.5 years; however, this estimator has a lower accuracy using the 450K BeadChip.39 Furthermore, it does not detect an association with mortality and revealed only a nominal correlation with TL or clinical and lifestyle measures (eg, alcohol consumption).
More accurate age predictions were reported with the rise in available data sets and advancements in technological/bioinformatic strategies. For example, Florath et al30 analysed blood DNA in 3 steps using a discovery (n = 965), replication (n = 400) and longitudinal cohort (67 individuals followed more than 8 years). It resulted in the selection of 17 CpGs to build a regression model for age prediction with an average error of only 2.6 years. However, the disadvantages of this DNAm clock are the use of a single tissue, the narrow age range of the cohort and the small number of selected CpGs (Table 1).
In contrast, Horvath11 multitissue clock consists of 353 CpGs and not significantly confounded by cell-/tissue-specific changes.5,40 It has been validated in multiple data sets, including ~8000 samples from 51 different cell and tissue types collected from both children and adults. Although the DNAm state of each CpG correlates only weakly with age, their combined effect results in an accurate biomarker of chronological age (an average error of 3-5 years). Additional measures of the Horvath clock include DNAm-age acceleration and intrinsic epigenetic age acceleration derived from adjusting DNAm-age for chronological age and blood cell proportions.41
The DNAm-age estimator of Hannum et al12 consists of 71 CpGs and is highly accurate in blood, but requires recalibration to achieve reasonable accuracy in other tissues. Both Hannum and Horvath clocks have a striking ability to predict all-cause mortality. They were built using a similar regression model for selecting age-related CpGs, and display moderate to strong correlations, even though they share only 6 CpGs.8 The Hannum clock and DNAm PhenoAge (discussed below) reflect age-related changes in cell and tissue-type composition.5 Notably, correcting for cell-/tissue-specific composition could eliminate biologically informative signals. For example, certain age-related diseases (eg, immunosenescence) are hallmarked by changes in the cellular structure of blood.5,42 Compared with intrinsic measures (consistent across cell/tissue types), extrinsic measures are more suitable for assessing age-related decline in specific tissues.5,41 DNAm-age acceleration measures are more informative, as they refer to discrepancies from the norm,5 and are associated with longevity and mortality giving the best representation of biological age.24,41,43
Zbiec-Piekarska et al32 analysed 41 CpGs by pyrosequencing of blood DNA (n = 420), of which 5 were included in an age predictor with an average error of 4.5 years. In a similar study, Huang et al31 analysed 38 different CpGs by pyrosequencing of blood DNA (n = 89), of which 5 were included in an age predictor with an average error of 7.9 years. In Zbiec-Piekarska clock, the mean absolute deviation varied from 5.0 for the oldest subgroup (age, 60-75) to 2.7 in the youngest subgroup (age, 2-19).32 The decreased accuracy of this clock in older people could reflect either technical issue(s) or the influence of external factors (eg, lifestyle). Several studies reported that DNAm changes slow down with age,6,12,13,44 which suggests the importance of including chronological age as a covariate. Furthermore, sex-specific differences were reported for Zbiec-Piekarska, Horvath and Hannum clocks, suggesting slightly faster aging in men than women, which could reflect either biological and lifestyle differences or technical issues affecting accuracy of the age predictors between the subgroups. Indeed, the mean absolute deviation of the Zbiec-Piekarska clock was marginally higher for men (3.7) than women (3.0).32
A recent large study by Zhang et al33 reported several DNAm predictors of chronological age based on the analysis of blood and saliva DNA. A near-perfect predictor of chronological age was developed using a training cohort covering a wide age range, which was further corrected for cellular composition. In nonblood tissues, it is comparable with the Horvath clock.11 However, the association between DNAm-age acceleration and mortality attenuates as prediction accuracy increases, which is likely the result of losing CpGs linked with biological age (during training overemphasizing the estimation of chronological age).
In summary, Horvath11 and Zhang33 clocks outperform other DNAm clocks based on either applicability to various cells/tissues or accurate prediction of chronological age, respectively. DNAm aging rate can be quite different between tissues,8 but clocks derived from a single tissue could be recalibrated to achieve a more accurate age prediction in another tissue.40 In general, tissue-specific clocks may better reflect age-related diseases,45 while multitissue clocks are more suited to capturing innate aging processes. Nevertheless, several studies reported associations between the acceleration of the Horvath multitissue clock and age-related disease risk or mortality, strongly supporting the potential of this clock as a disease biomarker.
.../...
.