By Vince Giuliano. With contributions, suggestions and editing by members of the YOUNGING team: Steve Buss, Walter Crompton, Chris Wikman and Debbie Coehlo
5-1-2022
GENERAL CONTEXT
From the onset some 12 years ago, this anti-agingfirewalls blog has followed an implicit Mission of contributing to people having and enjoying longer healthy and productive lives. Some of our posts have been discussions of practical and accessible interventions for doing this, such as taking certain dietary supplement,s or, more recently, exposing ourselves to PEMF interventions. Other posts have been on theoretical scientific frameworks for describing aging and anti-aging processes, characterizing the main theories described in the world’s scientific, literature. Many of the best practical interventions to secure health and long lives have been pursued for centuries or millennia with no understanding of how they work. But understanding them can lead to whole basic new frameworks of science. E.g. the drug rapamycin was discovered as something in the soil on Easter Island that extended the lives of small animals. It led to identification and details of the mTOR (Mammalian Target of Rapamycin pathway which conveys longevity when inhibited. A good theoretical framework can be very valuable for understanding interventions that work, and improving on them, and can lead to new interventions. The best paydirt is when we can couple a whole slew of well-known anti-aging interventions with a sound theoretical framework for understanding them. We have recently been successful in doing this, and decided that work using the term YOUNGING in multiple blog entries as well as Webcasts now available on YouTube.
YOUNGING Is our term for the innate restorative biological processes that go on in our bodies, like anti-inflammatory activity, epigenetic demethylation, wound healing, immune response, cell division, angiogenesis, stem cell replacement and DNA repair. There are a number of these processes that evolved as part of going from ephemeral, single-cell organisms, to extremely complex and highly regulated, long-lived creatures, with trillions of cells of many different kinds. Understanding YOUNGING mechanisms helps us understand known interventions to keep us as biologically young and healthy as long as is naturally possible. Understanding these mechanisms will also help us improve extant interventions and create new .
YOUNGING 1.0 describes YOUNGING processes that utilize extremely ancient, cellular, epigenetic (beyond genetics) histone demethylation approaches as well as more recent approaches that leverage the development of a Brain to Spleen mechanism for controlling inflammatory cytokine expression in the blood circulation.
Taken together, these two approaches make the activation of thousands of genes involved in YOUNGING growth and development possible.
So, please read the following blog entry in the context that it describes well-documented framework considerations that extend and enhance our earlier and narrower understanding of YOUNGING 1.0. Again, this is an enhancement to what we have published before related to YOUNGING 1.0and in no way denigrates or throws into question points made in those documents or presentations.
PURPOSE OF THIS BLOG ENTRY
The purpose of this blog entry is to convey a deeper understanding of global histone post-translational modification (PTM) phenomena to apply to our expanding understanding of YOUNGING. Specifically, this entry focuses on methyl- and acetyl- PTMs of H3K4 ,H3K9, H3K27, and bivalent PTM domains. These are complex topics, with my colleagues and I having only partial understanding to date. Grasping a rich new body of literature relating histone dynamics to natural restorative processes, aging, and strategies for age reversal will provide an important advance in our understanding of all aspects of YOUNGING. Further research in this area will help us understand how to trigger histone PTM processes and developmental or restorative in YOUNGING. Notably, this new area of knowledge fits with previous work on understanding YOUNGING, and I repeat that nothing we have published so far about YOUNGING is invalidated by this expanded view.
“Turning on” Genes
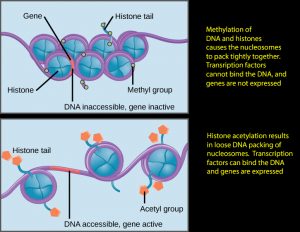
Previous understanding of the histone position stated, “Demethylation at histone position H3K27me2-3 by JMJD3 turns on genes near that position.” It is more accurate to say “Demethylation and/or of H3K27me2-3 opens up the neighboring chromatin, potentiating expression (transcription) of proximate genes.” (That is, it establishes a necessary condition for transcription of nearby genes to generate their associated proteins.)
Genes are NOT, however, automatically expressed due to demethylation or acetylation.. The genes’ promoter regions, and in some cases, co-promoters must also be activated for gene transcription to take place. A way to visualize this is that the bi/tri-methylation of H3K27me2-3 wraps the DNA into such tight knots that the promoter positions of the associated genes cannot be physically accessed by chemicals that would activate them. Demethylation by JMJD3 makes this access and transcription possible. So, you can think of the bi/trimethylation at H3K27me2-3 as a master lock that must be removed as a prerequisite of activation of nearby genes on Histone .
Unlocking this master lock via the histone demethylase JMJD3, we have asserted, is a NECESSARY condition for most if not all natural body restorative processes like stem cell renewal, expression of the OSKM factors or angiogenesis. It is also likely a prerequisite for most all YOUNGING intervention we can contrive to be workable. Being NECESSARY of course does not mean that induction of JMJD3 by itself is a SUFFICIENT condition to introduce general or specialized forms of YOUNGING.
More H3 histone trimethylation locations
Alas, this level of explanation is still overly simplified and ignores the roles of PTM at numerous other histone positions, including acetylation, SUMOylation, phosphoration, ribosilation and other factors that modify chromatin accessibility. I have focused recently on trimethylation of another two H3 positions: tri-methylation of histone H3 at the lysine located at position 9 (H3K9me3) and about tri-methylation of histone H3 at the lysine of position 4 (H3K4me3). And, histone bi and trimethylation atH3 position 79 (H3K79me2-3) These are different from our old friend H3K27me2-3, in that:
- different methyl transferases and demethylation substances are involved, and
- the forms of methylation have different effects on neighborhood gene activation. The trimethylation of H3K4me3 and H3K79me2-3results in open rather than closed chromatin (H3K4 is referred to as a “repressor” protein), chromatin in which gene activation is possible. Methylation at both H3K9me3 and H3K27me2-3 results in closed chromatin (I.e., they are “expressor” proteins); chromatin in which gene activation is impossible, So, H3K4me3 and H3K79me2-3are positive regulators of transcription and H3K9me3 & H3K27me3 are negative regulators (expressors) of it.
- Some genes are in regions of DNA neighboring two of these three H3 amino acid positions, that is positions 4, 9, 79 and 27. Such regions are called bivalent domains.
Further, there are methylation domains associated with H3K36 and with other histones in addition to H3 The situation can get maddeningly complex, so we need to take it on a little at a time, being always concerned for finding paydirt in effective health and longevity interventions. This blog is about being a little ways further down what could turn out to be a very long trail.
In summary, so far:
- Histone methylation at H3K27me2-3 is generated by operation of the polycomb repressive complexes (PRC1 & PRC2) and is removed by JMJD3 and UTX (another demethylase). – as laid out in earlier entries of this blog.
- Methylation at H3K9me3, in mammalian cells, can be catalyzed by at least six distinct SET domain enzymes: Suv39h1/Suv39h2, Eset1/Eset2 and G9a/Glp, and by operation of the PRCs. KDM4D demethylates di-and tri-methylation of histone H3 on the lysine at position 9. KDM4D is also known as JM
- Methylation at H3K4me3 happens by complex mechanisms (“Histone lysine methylation is generated by a battery of histone methyltransferases (HMTs) that transfer the methyl group from S-adenosylmethionine to specific lysine residues. For example, H3K4 methylation is mediated by several SET [Su(var)3-9, Enhancer of zeste, trithorax-group proteins] domain-containing methyltransferases, including mixed-lineage leukemia 1–5 (MLL1−5), SET1A/B, SET7/9, SET and MYND-domain-containing protein 1–3 (SMYD1−3), Absent, Small, or Homeotic1-like (ASH1L), SET domain and Mariner transposase fusion gene (SETMAR), and PR-domain zinc finger protein 9 (PRDM9) [5–24]. Methylated lysines exist in three forms: mono-, di- and tri-methylation (me1, me2, and me3.”) (ref). Histone methylation at H3K4me3 is removed by KDM5C/JARID1C.
- Histone methylation at H3K79me2-3: “— the yeast protein Dot1 and its human homolog, DOT1L, are responsible for catalyzing the methylation reaction (Feng et al. 2002; Lacoste et al. 2002; Ng et al. 2002a; van Leeuwen et al. 2002). Both enzymes are capable of catalyzing mono-, di-, and trimethylation in a nonprocessive manner (Min et al. 2003; Frederiks et al. 2008). Dot1 and its homologs appear to be solely responsible for H3K79 methylation, since knockout of Dot1 in yeast, flies, and mice results in complete loss of H3K79 methylation (van Leeuwen et al. 2002; Shanower et al. 2005; Jones et al. 2008).(reference)” “Here, we show that KDM2B, also known as FBXL10 and a member of the Jumonji C family of proteins known for its histone H3K36 demethylase activity, is a di- and trimethyl H3K79 demethylase.(reference)
Bivalent domains
We have thus learned that there are more H3 trimethylation mechanisms to enable or disable masses of genes to be switched on or off than just the PRC/JMJD3 combo. This gives cells much greater and finer control for enabling or disabling large groups of genes. It can do this by using different histone methylases/demethylases. Evolution no doubt put these mechanisms in place to facilitate turning expressibility of whole subsets of developmental genes off and on in the complex processes of organ and structure development in complex organisms like our bodies, In many cases of natural adult renewal of body parts or systems, like stem cell differentiation, the same developmental genes are naturally invoked again and again. And the same steps of histone methylation and demethylation are necessary to allow or inhibit expression of those genes.
There is; however, a rub. Some genes are located on nucleosome spindles so they are affected simultaneously by a PAIR of these methylated histone positions. Such locations are called bivalent domains. What happens to genes in these bivalent domains, say when if both H3K4me3 and H3K27me3 are methylated, the former leading to open chromatin, the second leading to closed chromatin? Again, there is a lot of research on this question and its implications which can get really complicated and involuted. The bottom line is that the body uses these bivalent domains as epigenetic switches which figure significantly in original organism development, in natural regenerative processes, and almost certainly they will figure heavily in any effective YOUNGING interventions we can contrive.
View of some possible post-translation modifications associated with one histone. On H3 on lower left, count over to position 27. K stands for lysine. Only acetylation and methylation are show as possible modifications. Also, only the first 38 positions are illustrated. Image source Wikipedia.
The methylation and de-methylation of the three different H3 positions discussed depend on different methylases and different demethylases. So, our bodies body can control their methylation status independently. This fact allows bivalent domains to act as powerful epigenetic switches for opening up or closing in the chromatin in them. That is; the bivalent domains consist of sequences of genes that can all at once be switched from being transcribable or not transcribable, or the other way. Consider this Table for a bivalent domain influenced by methylation-versus-acetylation both at H3, positions 4 (a repressor) and 27 (an expressor), for example, where ’me’ means the methylation is present, and ‘ac’ means acetylation is present. So, by toggling between the situations described in B and C in the Table all the genes in the bivalent domain can be made accessible for transcription, or all can be made not accessible
| H3K27me3 | H3K27ac | |
| H3K4me3 | Mixed chromatin. Some genes in domain expressible A | Open chromatin. Genes in domain expressible B |
| H3K4ac | Closed chromatin. Genes in domain not expressible C | Mixed chromatin. Some (other) genes in domain expressible D |
Many promoter locations on embryonic stem cells are influenced by this particular bivalent domain situation. Bivalent domains are thus like wall toggle light switches, which allow easy switching back and forth to inducing stable open or stable closed chromatin, as illustrated in this diagram. They are what allow stem cells to have alternative fates.
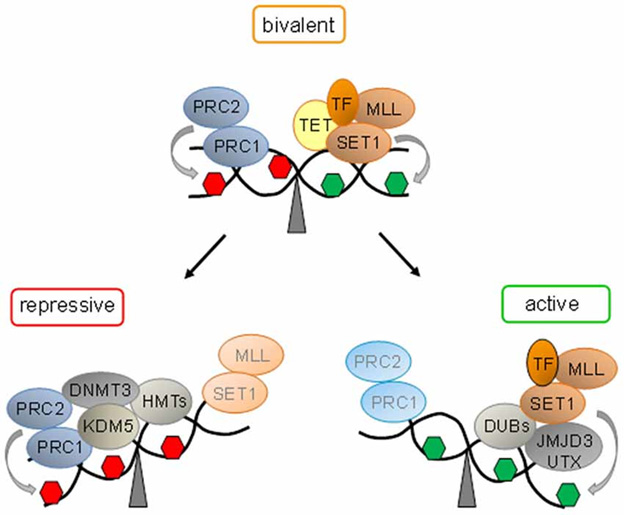
“At bivalent domains, the simultaneous presence of repressive (red hexagon) and active (green hexagon) PTMs counterbalance each other. PRC1 and PRC2 confer repression by H3K27me3 catalyzed by the subunit Suz12. Ten − eleven translocation proteins (Tet) interact with bivalent domains and catalyze DNA demethylation. Activating transcription factors (TF) together with histone H3K27 demethylases (SET1 and MLL) and H2A-deubiqitinating enzymes (DUBs) tilt the balance towards activation, and replacement of repressive protein complexes. Conversely, loss of activating protein complexes switches bivalent domains into a repressed state by recruitment of H3K4 demethylase (KDM5). Histone methyltransferases (HMT) mediated H3K9 methylation and de novo DNA methyltransferase (DNMT3) mediated DNA methylation lock in repression.” Source of diagram and legend: Bivalent domains underlie epigenetic switches in stem cells
The following illustration shows how a bivalent domain of particular interest to us can be formed:
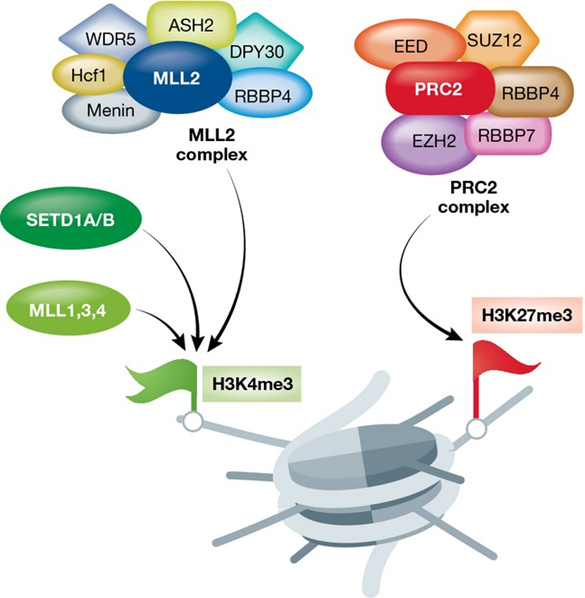
Activation of a bivalent domain “Left: Protein complexes catalyzing H3K4 methylation (green flag). Right: The PRC2 complex catalyzing H3K27 methylation (red flag). Shown are only the main proteins and protein complexes catalyzing H3K4/H3K27 methylation. Less abundant subunits are not depicted. [Correction added on 2 December 2015 after first online publication: “H4K4” has been corrected to “H3K4”.]” Source: Chromatin remodeling and bivalent histone modifications in embryonic stem cells (2015)
This publication describes one of the steps where bivalent domains are involved in creating mammalian life: Polycomb protein SCML2 facilitates H3K27me3 to establish bivalent domains in the male germline “Our study identifies a possible germline mechanism by which differentiated and unipotent germ cells give rise to a totipotent zygote following fertilization.”
Difficulty of ChiP-seq identifying true bivalent domains
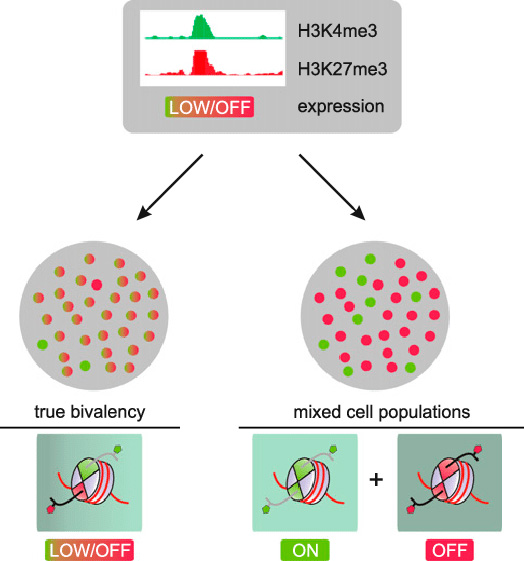
The following image refers to the same bivalent domain bordered by H3K4 and H3K27. The point being made is that ChIP-seq measurements may show both H3K4me3 and H3K27me3, but these may or may not be originated in the same cell, so this may or may not represent the presence of bivalent domains.
Image source “Bivalent domains and heterogeneity. Two scenarios could potentially explain the co-occurrence of H3K4me3 and H3K27me3 observed by ChIP-seq on bivalent promoters. As ChIP-seq cannot establish physical co-occurrence of two marks on the same allele, admixture of cells that either express (green) or do not express (red) the gene in focus could explain the occurrence of both marks as well as the low expression level in the overall population. In contrast, in the case of ”true” bivalency, virtually all cells in the population carry both marks simultaneously at the promoter in question, leading to low, if any, expression for that gene in all cells.”
Developmental and restorative processes depend crucially on the operations of bivalent domains
Specifically, cell reproduction and stem cell operations depend on bivalent domains as explained in the 2021 publication Histone Lysine Methylation and Long Non-Coding RNA: The New Target Players in Skeletal Muscle Cell Regeneration “Transcriptional regulators, including Nanog, Oct4, and Sox2 that maintain the pluripotency of stem cells, must be deactivated for cellular differentiation to occur (Wang et al., 2012; Chu et al., 2016). In addition to transcriptional regulators, induced pluripotent stem cells (iPCS) and embryonic stem cells (ESC) are poised by a bivalent chromatin, consisting of histone 3 lysine 4-trimethylation (H3K4me3) and histone 3 lysine 27-trimethylation (H3K27me3) (Bernstein et al., 2006; Mikkelsen et al., 2007). These bivalent marks are located on the same nucleosome in an asymmetric configuration regulating the chromatin structure (Bernstein et al., 2006). Generally, H3K4me3 is associated with gene activation, while H3K9me3 and H3K27me3 are associated with gene repression (Black et al., 2012). Upon receiving differentiation cues, the H3K27me3 repressive mark is lost, and H3K4me3 activity dominates the promoter region lineage-specific genes to activate gene transcription (Collinson et al., 2016). The bivalent gene is essential to regulate the transition between pluripotency and committed cells. Moreover, the H3K27me3 repressive mark of the bivalent gene maintains low expression levels of developmental genes in iPCS and ESC while allowing for their transcription upon differentiation. Furthermore, H3K27me3 protects the cell from aberrant gene activation by permitting only the target lineage-regulating genes to activate (Vastenhouw and Schier, 2012). H3K4me3 has vital significance as a transcriptional activator. In addition to this main function, H3K4me3 may be essential to ensure that permanent gene silencing of developmental genes does not occur (Fouse et al., 2008; Vastenhouw and Schier, 2012).”
Chromatin Remodelers
There are certain “chromatin remodeler” proteins that work by affecting bivalent domains. For example, the following chart and diagram are from Chromatin remodeling and bivalent histone modifications in embryonic stem cells (2015). That document goes into a great deal of detail about processes of generation and destruction of bivalent domains and their effects on chromatin. It is a good place to look if you wish to dig deeper.
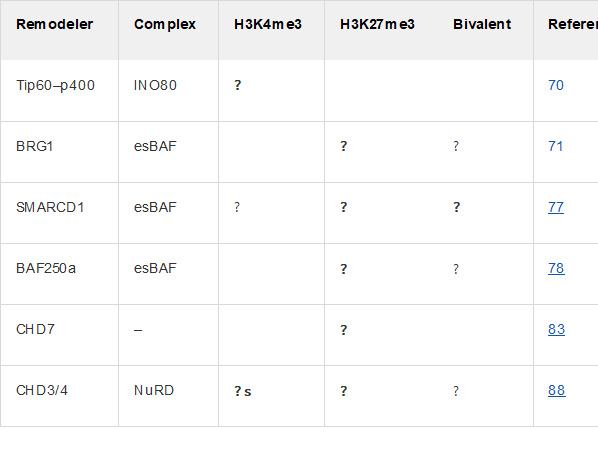
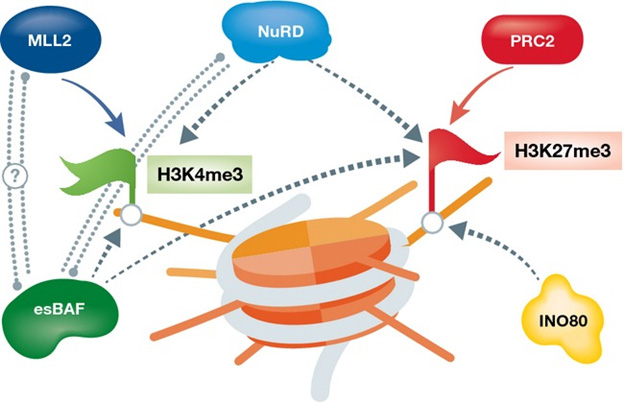
ChromChromatin remodeling complexes regulating bivalent nucleosomes “A single schematic bivalent nucleosome is shown (orange) marked with both H3K4me3 (green flag, left) and H3K27me3 (red flag, right). Chromatin remodeling complexes which were shown to regulate either or both marks are shown in green (esBAF), blue (NuRD), and mustard (INO80). Dotted arrows represent suggested regulation; dotted double lines represent potential interaction.”
SOME KEY OBSERVATIONS
Epigenetic regulation by bivalent domains is extremely ancient and applies to most living species, including mushrooms.
Heterochromatin regulation via methylation of these histone goes back over 400 million years of evolutionary history and applies widely across very diverse living species. So, for example, experiments on an ancient mushroom species may be directed towards finding insights relevant to humans today. See for example the paper Induction of H3K9me3 and DNA methylation by tethered heterochromatin factors in Neurospora crassa. The Histone Code provides a systematic framework for biology as does the periodic Table for Chemistry.
Potatoes also employ essentially the same bivalent domains as we do for epigenetic regulation.
The 2019 publication Cold stress induces enhanced chromatin accessibility and bivalent histone modifications H3K4me3 and H3K27me3 of active genes in potato makes this point. Mushrooms, potatoes and people? The same mechanism also applies to flies, nematodes(ref) and all mammals. And oh yes, the vemon of tarantulas and snakes depend on bivalent domains(ref). This blog entry is talking about very basic tried-and-true mechanisms of biology.
The key roles of histone regulation in biology were discovered not that long ago, and their implications have not yet been appreciated in many areas of the science, including longevity science
Up to this point I have seen no mention in the longevity-related literature about bivalent domains, nor do I know of any longevity science researcher who have expressed any care about them. Yet, just scratching the surface I already see much evidence that bivalent domains are basic to natural YOUNGING processes. They will no doubt figure heavily jn any YOUNGING interventions that work. This timetable is from a 2012 publication and could be expanded considerably to reflect developments in the last10 years. Image and legend source.
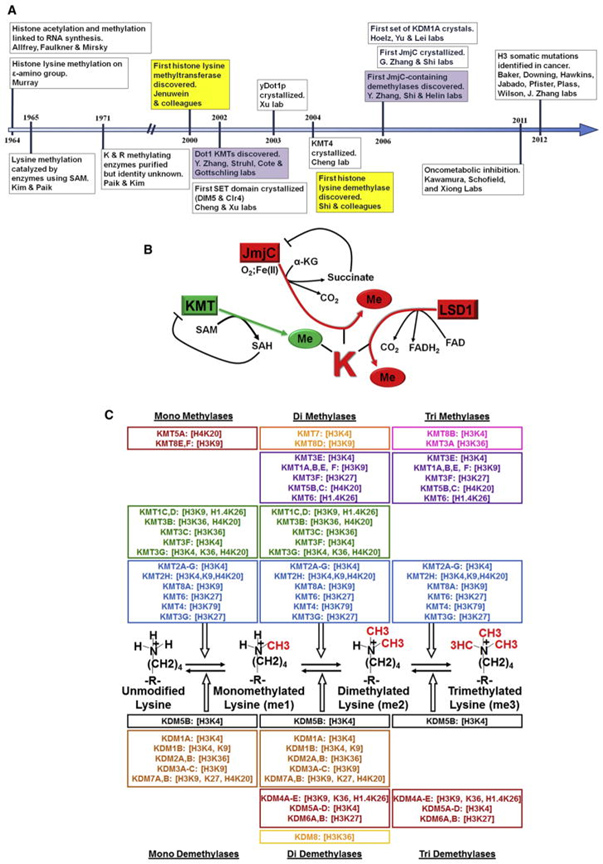
History, Mechanism, and Specificity of KMTs and KDMs
(A) Timeline chronicling important milestones in KMT and KDM research. (B) Schematic depicting generalized reaction mechanisms of KMTs and KDMs. © Schematic depicting substrate specificity of KMTs and KDMs”
THESE TOPICS ARE VERY RELEVANT FOR OUR EXPLORATION OF YOUNGING AND YOUNGING INTERRVENTIONS
For one matter they can point us to vast new chapters of the biomedical literature relevant to YOUNGING, with articles like these:
Bivalent-histone-marked immediate-early gene regulation is vital for VEGF-responsive angiogenesis
Complete loss of H3K9 methylation dissolves mouse heterochromatin organization
Synergistic lethality between BRCA1 and H3K9me2 loss reflects satellite derepression
Repressive Chromatin in Caenorhabditis elegans: Establishment, Composition, and Function.
H3K9 methyltransferase G9a and the related molecule GLP.
H3K9me selectively blocks transcription factor activity and ensures differentiated tissue integrity.
The Sound of Silence: How Silenced Chromatin Orchestrates the Repair of Double-Strand Breaks.
Bivalent Epigenetic Control of Oncofetal Gene Expression in Cancer.
Bivalent Histone Modifications and Development.
ON GOING DEEPER INTO THE HISTONE CODE
The material covered here represents highly selected aspects of the Histone Code, which is concerned with the impacts on gene expression of all histone modifications on all histones, (that is modifications due to histone methylation, acetylation, ribosylation, ubiquitination, threonine/serine/tyrosine phosphorylation, etc.); considering not only individual modifications but viable combinations of them. The complexity is mind-numbing. Wikipedia points out “Unlike this simplified model, any real histone code has the potential to be massively complex; each of the four standard histones can be simultaneously modified at multiple different sites with multiple different modifications. To give an idea of this complexity, histone H3 contains nineteen lysines known to be methylated—each can be un-, mono-, di- or tri-methylated. If modifications are independent, this allows a potential 419 or 280 billion different lysine methylation patterns, far more than the maximum number of histones in a human genome (6.4 Gb / ~150 bp = ~44 million histones if they are very tightly packed). …And this does not include lysine acetylation (known for H3 at nine residues), arginine methylation (known for H3 at three residues) or threonine/serine/tyrosine phosphorylation (known for H3 at eight residues), not to mention modifications of other histones. – Every nucleosome in a cell can therefore have a different set of modifications, raising the question of whether common patterns of histone modifications exist. A study of about 40 histone modifications across human gene promoters found over 4000 different combinations used, over 3000 occurring at only a single promoter. However, patterns were discovered including a set of 17 histone modifications that are present together at over 3000 genes.[16] Therefore, patterns of histone modifications do occur but they are very intricate, and we currently have detailed biochemical understanding of the importance of a relatively small number of modifications.”
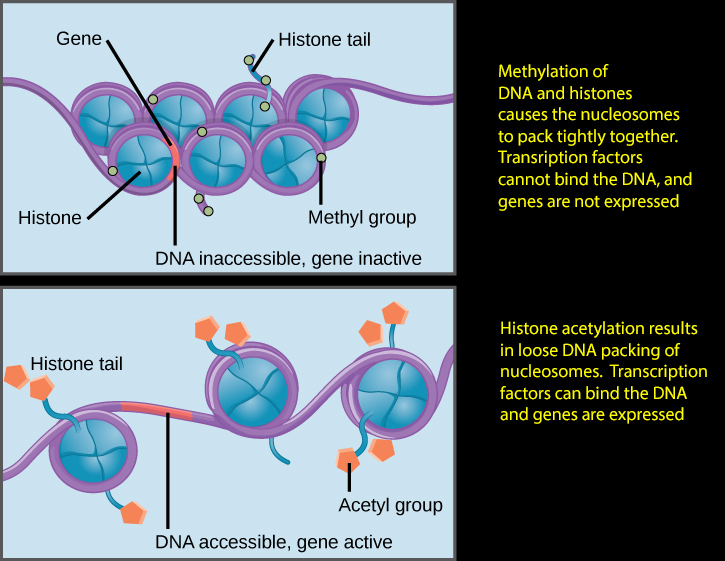
Note that histone methylation compacts the chromatin as shown only for certain domains like H3K27, the opposite being true for others like H3K4
Also mind-numbing is the likely payoff of deeper understanding of the histone code for helping us move down the road to clarifying optimal science-based longevity interventions. This blog entry talks about only methylation modifications affecting only histone 3, so it provides only a start. We need learn more aspects of this code as we go on, perhaps as the next step on the combined impacts of histone methylation and acetylation, given that we know a fair amount about many natural substances can be powerful histone acetylases/deacetylases. They offer powerful natural approaches to activating key longevity pathways including SIRT1 andSIRT6 and the positive effects of enhancing NAD. See several articles in this blog by myself and by Jim Watson, starting with my Histone acetylase and deacetylase inhibitors post. Also, Jim Watson’s PART 2: Slaying Two Dragons with One Hail of Stones: The Silencing Of Good Genes In Aging And Cancer – And How Polyphenols Can Prevent That. So, I expect there will be a lot more to come from me and my YOUNGING colleagues about YOUNGING and the Histone Code.
————–———————————————
Doe
View the full article at Anti-Aging Firewalls