| By Vince Giuliano, inspired by interaction by James P. WatsonThis is the first blog entry in a three-part series concerned with transposable DNA entries (TEs). It focuses on basics of what TEs are, why they are important for us who are concerned with health and aging, and briefly outlines what comes later in the series. Part 2 of the series, The Self-copy Machines in Your Genes – transposable elements was mainly generated by Jim Watson. It is the main course of the series, providing the “meat and potatoes” of technical content. Twenty one key aspects of TEs are discussed there. Part 3 of the series TEs and an emerging “Grand Unified Theory” of biology and aging outlines the key role of TEs in making evolution possible in response to a changing environment, and how they fit into an emerging unified theory of how all life and aging works.What are transposable elements?Most of the DNA in our human genome is composed of largely but not completely, inactive copies of three distinct classes of transposable element, sequences of DNA that have accumulated over millions of years and keep replicating. “A transposable element (TE or transposon) is a DNA sequence that can change its position within the genome, sometimes creating or reversing mutations and altering the cell’s genome size. Transposition often results in duplication of the TE(Wikipedia).” A transposable element is a segment of DNA with inverted terminal repeats at each end that can excise or copy itself and move from one location to another within or between chromosomes, plasmids, or phage. Not only can some TEs copy their own genomes — they can also copy and paste parts or wholes of adjacent human genes. And they can relocate or paste in the DNA where they want, including in the middle of other genes. Although we are still starting to understand this, we know it is not completely random.TEs have played enormously important roles in shaping our evolution and some of these TEs are continuing to do that today. Transposable elements have been called “jumping jeans.” Most of these TEs are in fact not protein-coding genes, but jump around they do. They also may be a major driver of aging. Key practical facts about transposable elements Despite the fact that there are different classes of TEs with different properties, a few important generalizations can be drawn, including: TEs and evolution TEs seem active in all organisms and to be major drivers of evolution within and between species. Tracing our evolution, some of the first TEs showed up over 150 million years ago in primitive organisms like yeast. As time progressed, we can identify the emergence of TE classes of increasing sophistication. Alu is a primate-specific transposable element whose copy numbers have increased from zero to between 1.4 and 2 million copies over the past 65-80 million years of primate evolution. Some recently emerged TEs seem to apply only to us hominids, like SVAs which go back only 25 million years or so. TEs can systematically broadcast complex genetic mutations in DNA under stress conditions. They can copy or export long segments of DNA to remote and seemingly unrelated places in the genome of a cell. It is as if they can say to the organism concerned “It seems like you are experiencing a lot of stress and I am getting concerned about your survival and success as a species. So, I am going to throw your evolution machine into higher gear by mixing up your DNA a bit, copying whole pieces and moving them to new places in your genome. Sometimes, I will make you another copy of one of your genes in a new place. Or, even several copies. Some of those possible DNA shifts or mutations may give you cancers or kill you or otherwise screw you or your progeny up. I am sorry for that, but I hope you understand that my deep commitment is to the evolution and proliferation of species, not to you as an individual, or even to your particular species. That means innovation in the interest of life going on and developing life further as conditions change. And sometimes it means creating new species. These in turn require innovation in your genome and that is my job. However, I generally don’t generally do random mutations. Mostly, the pieces of DNA I spread around or duplicate in the genome of one of your cells are ones you had somewhere there in the first place. Many of the innovative DNA shifts and mutations in your genome, perhaps in other individuals like you, will prove useful and enhance survival or perhaps emergence of a superior species. I know this works well since we, the TEs, have been doing this for millions of years and it has been key to your species of homo sapiens getting to where it is now. Years ago you learned that evolution was due to random variation and natural selection. What I am telling you now is that the variation is deliberate, is triggered, and focused. Speaking on behalf of me and my fellow TEs, I am telling you that generating that variation is our job.” In other words, TEs are a very deep mechanism for rapid evolution, applying across species, going back to near the beginning of life forms. If you like the idea of being more than a slime mold, you should appreciate them. The Part 3 blog entry in this series further discusses the role of TEs in evolution. TEs and health Most TEs in humans are no longer active although still present in our DNA and making up about half of our genome. The vast majority of these are no longer active as a result of cumulative mutation, truncation, internal rearrangement and silencing. However, it is estimated that approximately 100 of L1s TEs per nuclear genome still retain their replication activity. If promoted by cancer, certain other disease conditions and aging, these can create mutations that lead to pathologies and diseases. Some Line-1 TEs can create single and double-strand DNA breaks. Alu TE element copying can introduce insertions and deletions in 33 genes, leading to many more terrible diseases. SVA TEs can exercise DNA mischief including the generation of differentially methylated regions, exon shuffling, alternative splicing, and of course insertional mutations. Some TEs can do strange things in terms of gene activation. For example, an L1 TE may contain an antisense promoter leading it to activate an unrelated upstream gene, or, more likely, down-regulate it or silence it. TEs of the Alu and LTR families can provide binding sites for NF-kappaB, master regulator of immune response and inflammation. So, TEs can turn on inflammation. Cancer cells and certain other diseased cells have very high levels of TE expression, especially cancer cells that have progressed to late stages of chemotherapy and radiation resistance. TEs may be implicated in Alzheimer’s disease through creating many extra copies of the gene that makes amyloid precursor protein in the cortical neurons of AD patients. Human endogenous retroviruses (HERVs) are TEs, but though they make up a great deal of our genome they are largely inactive as viruses. However, they are quite important since their long terminal repeats (LTRs) can serve as alternative promoters of nearby genes. This appears to be a process very important for embryogenesis, but can also lead to a number of disease conditions. TEs and aging TEs are highly likely to play major roles in aging. There is widespread agreement that genomic instability and DNA damage are key features of aging – and TEs cause both. Although the research on this topic is not completely definitive yet, it may well turn out to be the case that TEs are more important for aging in elderly people than the “usual suspect” pathways like IGF-1 and mTOR. TEs are likely to do this by generating double strand DNA breaks, by generating insertional mutations, by silencing of genes, by gene copy number increases, by triggering pathways that lead to cellular senescence and apoptosis, and by generating inflammation through activating alternative binding sites for inflammatory factors. Global hypo-methylation associated with aging leads to activation of TEs. Consistent with this, the expression of certain TEs, namely LINE-1 elements, dramatically increases with aging in yeast, in fruit fly brains, in mouse liver and skeletal muscle, and in human somatic cells. A few studies have shown that overexpression of L1 can cause cells to senesce. Some researchers think that large numbers of DNA breaks associated with L1 TEs may be a major factor in aging. Such breaks can exhaust limited supplies of NAD+ and SIRT1 for their repair, impairing mitochondrial and metabolic health. expression of L1 TEs can induce cellular senescence or apoptosis in normal cells. Studies of SNPs of genes that are promoted by LTRs in TEs suggest an intriguing link between inflammation and aging. When SNPs of inflammation-promoting genes are present, they cannot be activated by TEs and longer lifespans result. TEs are responsible for increasingly mixing up our non-germline DNA as we age, leading us to become “genomic mosaics” where not all of the cells have the same copy number of genes and some cells may have multiple copies of pro-aging genes. What activates TEs and what silence them? No surprise, inflammation and major stresses are the big activators if TEs. Some TEs can turn on inflammation – human endogenous retroviruses (HERVs) and LTR remnants of HERVs can turn on inflammation by using the LTRs as “alternative promoters” for NF-kB and many other inflammatory transcription factors. In turn, inflammation itself can activate TEs. There is now strong evidence that transposable elements become activated and TE transcription increases with inflammation and cellular stress. The consequences can be serious, such as in the case of inflammation induced by IFN-gamma. Some of the inflammatory transcription factors produced by IFN-gamma can activate LTRs on TEs found near human genes. These inflammatory transcription factors can use the LTR as a “landing strip” and activate nearby genes that can induce cancer and aging. This is why chronic viral infections are associated with an increased risk of cancer and accelerated aging. As to keeping TEs silent, we humans are much better at this than lower animals, and this might be a big part of explaining why we live longer. However, for the reasons outlined above, implementing measures for keeping TEs silent as we become aged can be very important. Fortunately, there seems to be a number of strategies for doing this. As just indicated, keeping inflammation down is an excellent idea – a recurrent theme from a number of other longevity-related viewpoints. Some of our dear old anti-aging friends like SIRT6, melatonin, caloric restriction and fasting can be helpful in keeping TEs silent. Keeping our level of NAD and our NAD/NADH ratio up can be helpful for having enough SIRT6 around to both handle double-strand DNA breaks and keep our L1 TEs silent. Fasting and other measures that down-regulate the insulin/IGF-1 pathway might help. Also, lifestyles that respect our circadian rhythms, biotin, histone deacetylase inhibitors, histone acetyl transferase inhibitors, and sirtuin activators may also be promising approaches to suppress TE expression. And vaccines might help, ones being developed to work against ORF2 and HERV proteins for preventing cancer and HIV infections. Finally, some drugs might turn out to be useful in this regard, such as reverse transcriptase inhibitors like lamivudine and adefovir. For the research and literature citations backing up the above statements, see Jim Watson’s Part 2 blog entry in this series.
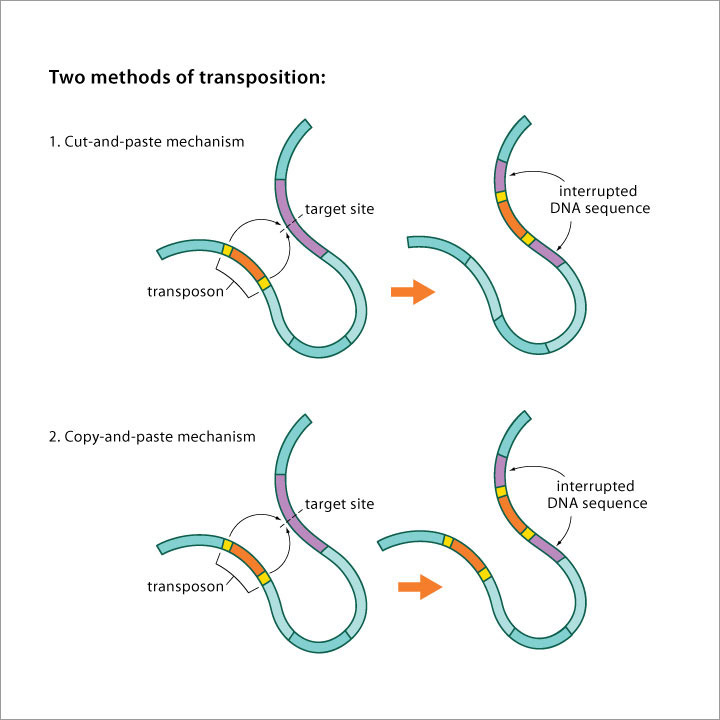
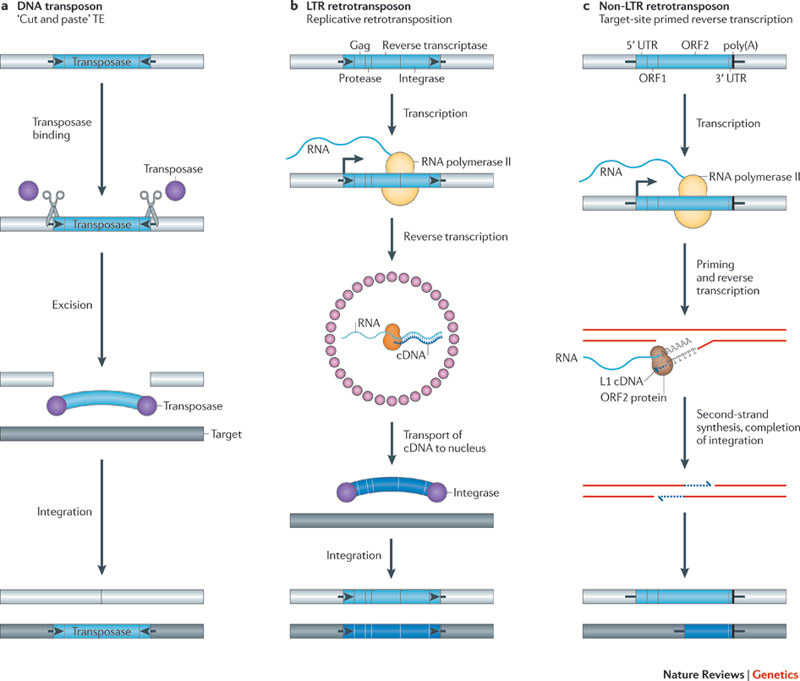
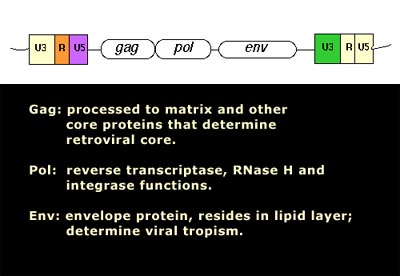
Image source Genetic mosaicism in corn due to transposable elements Background homework on TEs If you are not familiar with the basics of transposable DNA elements (TEs), I suggest you start by reviewing some basic information about them, such as on the Wikipedia page on this topic. I have quoted a few passages from that page here in italics to introduce a number of distinctions that are referred to throughout this blog entry. You can skip this Section and go right on to the Part 2 blog entry if you already are familiar with TEs. · Again from Wikipedia: “Transposable elements (TEs) represent one of several types of mobile genetic elements. TEs are assigned to one of two classes according to their mechanism of transposition, which can be described as either copy and paste (class I TEs) or cut and paste (class II TEs).[12]” “Class I (retrotransposons) Class I TEs are copied in two stages: first they are transcribed from DNA to RNA, and the RNA produced is then reverse transcribed to DNA. This copied DNA is then inserted at a new position into the genome. The reverse transcription step is catalyzed by a reverse transcriptase, which is often encoded by the TE itself. The characteristics of retrotransposons are similar to retroviruses, such as HIV. Retrotransposons are commonly grouped into three main orders: · TEs with long terminal repeats (LTRs): encode reverse transcriptase, similar to retroviruses · LINEs (LINE-1s or L1s): encode reverse transcriptase, lack LTRs, and are transcribed by RNA polymerase II · SINEs: do not encode reverse transcriptase and are transcribed by RNA polymerase III. Retroviruses can also be considered TEs. For example, after entering a host cell and conversion of the retroviral RNA into DNA, the newly produced retroviral DNA is integrated into the genome of the host cell. These integrated DNAs represent a provirus of the retrovirus. The provirus is a specialized form of eukaryotic retrotransposon, which can produce RNA intermediates that may leave the host cell and infect other cells. The transposition cycle of retroviruses has similarities to that of prokaryotic TEs, suggesting a distant relationship between these two TEs types. Class II (DNA transposons) The cut-and-paste transposition mechanism of class II TEs does not involve an RNA intermediate. The transpositions are catalyzed by several transposase enzymes. Some transposases non-specifically bind to any target site in DNA, whereas others bind to specific DNA sequence targets. The transposase makes a staggered cut at the target site resulting in single-strand 5′ or 3′ DNA overhangs (sticky ends). This step cuts out the DNA transposon, which is then ligated into a new target site; this process involves activity of a DNA polymerase that fills in gaps and of a DNA ligase that closes the sugar-phosphate backbone.[citation needed] This results in duplication of the target site. The insertion sites of DNA transposons may be identified by short direct repeats (created by the staggered cut in the target DNA and filling in by DNA polymerase) followed by a series of inverted repeats important for the TE excision by transposase. Cut-and-paste TEs may be duplicated if their transposition takes place during S phase of the cell cycle when a donor site has already been replicated, but a target site has not yet been replicated.[citation needed] Such duplications at the target site can result in gene duplication, which plays an important role in evolution.[13]:284 Not all DNA transposons transpose through the cut-and-paste mechanism. In some cases, a replicative transposition is observed in which a transposon replicates itself to a new target site (e.g. Helitron (biology)). Class II TEs make less than 2% of the human genome, making the rest Class I.[14] Autonomous and non-autonomous TEs Transposition can be classified as either “autonomous” or “non-autonomous” in both Class I and Class II TEs. Autonomous TEs can move by themselves while non-autonomous TEs require the presence of another TE to move. This is often because non-autonomous TEs lack transposase (for class II) or reverse transcriptase (for class I).” · The various kinds of transposons are mobile genetic elements that can move about in the genome, and there are other such elements as well including Plasmids, Bacteriophage elements, like Mu, Group II introns and Group I introns. The total of all mobile genetic elements in a genome may be referred to as the mobilome. This diagram illustrates the cut-and-paste and copy-and-paste mehanisms of movement. Here is a somewhat more complete diagram showing what TEs do: Image source: 2011 Dynamic interactions between transposable elements and their hosts Fig 1 Additional key distinctions ORF -open reading frame” In molecular genetics, an open reading frame (ORF) is the part of a reading frame that has the potential to code for a protein or peptide. An ORF is a continuous stretch of codons beginning with a start codon (usually AUG) and ending with a stop codon(usually TAA, TAG or TGA).[1] ” (ref) LTR – long terminal repeat. Long terminal repeats (LTRs) are identical sequences of DNA that repeat hundreds or thousands of times found at either end of retrotransposons or proviral DNA formed by reverse transcription of retroviral RNA. They are used by viruses to insert their genetic material into the host genomes. RT– Reverse transcriptase (RT) is an enzyme used to generate complementary DNA (cDNA) from an RNA template, a process termed reverse transcription ALU – An Alu element is a short stretch of DNA originally characterized by the action of the Alu (Arthrobacter luteus) restriction endonuclease.[1] Alu elements of different kinds occur in large numbers in primate genomes. In fact, Alu elements are the most abundant transposable elements in the human genome. They are derived from the small cytoplasmic 7SL RNA, a component of the signal recognition particle. The event, when a copy of the 7SL RNA became a precursor of the Alu elements, took place in the genome of an ancestor of Supraprimates.[2] HERVs – Human endogenous retroviruses Gag, Pol and Env – these are three major proteins encoded in the retroviral genome:
Image and legend reference ”The group antigens form the viral core structure, RNA genome binding proteins, and are the major proteins comprising the nucleoprotein core particle. Reverse transcriptase is the essential enzyme that carries out the reverse transcription process that take the RNA genome to a double-stranded DNA preintegrate form. — The reverse transcriptase gene also encodes an Integrase activity and an RNase H activity that functions during genome reverse transcription.” If you are intrigued by TEs in the light of the above, I strongly suggest you proceed to Jim Watson’s part 2 blog entry. The 21 items discussed there are: 1. There are Over 2 Million Copy Machines in your Genes, but only 80-100 still work (they are all LINE-1s) 2. Short-lived organisms are more permissive to retrotransposition, whereas long-lived organisms are resistant to retrotranspostion. 3. Human Gene Transcription or Repression can be triggered from the anti-sense promoters of LINE-1 and from the bidirectional LTRs of old HERV sequences 4. TEs and inflammation 5. Bidirectional promoters and diseases 6. SIRT6 “vacates” L1 promoters with aging, allowing L1 expression to occur in old age 7. Inflammation triggers transposable element expression 8. Germ lines and stem cells are protected against TE expression 9. Knocking out the Insulin/IGF pathway in C. elegans silences transposons by RNAi, Piwi-piRNA pathway, and by TDP-1 10. Transposable element endonuclease (ORF2) induces double stranded DNA breaks (DSBs) via endonuclease “nicking” of DNA 11. Cancer is manifested by dramatic increases in transposable element expression 12. LTR transposable elements contain G-quadruplex sequences at specific distances from their endogenous TE promoter that may regulate LTR-mediated gene expression 13. Transient LINE-1 over-expression induces cell senescence or apoptosis in normal healthy cells 14. Melatonin suppresses LINE-1 expression, as does sleep and turning off the lights at nigh 15. SIRT6 – An important silencer of transposable element expression and a candidate “longevity gene” 16. Longevity is associated with gene polymorphisms that may play a role in reducing TE activity and reducing inflammation 17. The human brain is a very active site of L1 transcription and insertions into neurons, in a cell-specific manner that produces gene mosaicism 18. TE-originated genetic mosaicism – a likely cause of Alzeimer’s disease 19. Vaccines with cancer-associated ERVS and HIV infection-associated ERVs appear to be safe and immunogenic 20. Reverse Transcriptase inhibitors that suppress transposable element retrotransposition may effectively treat cancer 21. Retrotransposon activity is likely to be a MAJOR cause of aging |
View the full article at Anti-Aging Firewalls